Abstract
The structure of the title compound, [NaRuCl4(C2H6OS)2]n, comprises centrosymmetric [RuCl2(DMSO)Na(DMSO)Cl2Ru] units (DMSO is dimethyl sulfoxide, C2H6OS), with two Ru atoms, each lying on a crystallographic centre of inversion, connected via Na atoms, DMSO and chloride ligands into a two-dimensional (110) array. Both RuIII atoms are octahedrally coordinated by four chloride ligands in the equatorial plane and by two DMSO molecules in apical positions within a RuCl4S2 donor set. The Na atom is surrounded by three chloride anions and three O atoms derived from three DMSO molecules, with the resulting Cl3O3 donor set defining an octahedron. The crystal structure is further stabilized by interatomic interactions of the types C⋯Cl [C—Cl = 3.284 (2) Å], C—H⋯Cl [C⋯Cl = 3.903 (3) Å] and C—H⋯O [C⋯O = 3.376 (3) Å].
Related literature
For structures of similar ruthenium complexes, see: Alessio et al. (1993 ▶); Piggot et al. (2004 ▶); Anderson et al. (2007 ▶). For Na—O and Na—Cl distances in related structures, see: Alessio et al. (1991 ▶); Iengo et al. (1999 ▶).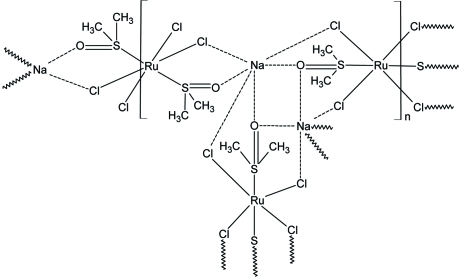
Experimental
Crystal data
[NaRuCl4(C2H6OS)2]
M r = 422.12
Monoclinic,
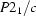
a = 11.9042 (3) Å
b = 8.0692 (2) Å
c = 13.7873 (3) Å
β = 98.470 (2)°
V = 1309.93 (5) Å3
Z = 4
Mo Kα radiation
μ = 2.34 mm−1
T = 120 K
0.20 × 0.20 × 0.15 mm
Data collection
Oxford Diffraction Xcalibur2 CCD diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007 ▶) T min = 0.652, T max = 0.721
10197 measured reflections
2300 independent reflections
2105 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.018
wR(F 2) = 0.050
S = 1.15
2300 reflections
134 parameters
H-atom parameters constrained
Δρmax = 0.71 e Å−3
Δρmin = −0.36 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2007 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2007 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810007063/tk2632sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810007063/tk2632Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Ru1—S1 | 2.3350 (5) |
| Ru1—Cl1 | 2.3509 (5) |
| Ru1—Cl2 | 2.3551 (5) |
| Na1—O2 | 2.2974 (18) |
| Na1—O1 | 2.4105 (17) |
| Na1—O1i | 2.4155 (18) |
| Na1—Cl4ii | 2.7773 (11) |
| Na1—Cl1iii | 2.8769 (10) |
| Na1—Cl2iv | 2.9374 (10) |
| Ru2—Cl3 | 2.3353 (6) |
| Ru2—S2 | 2.3373 (6) |
| Ru2—Cl4 | 2.3663 (6) |
Symmetry codes: (i) 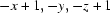 ; (ii)
; (ii) 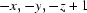 ; (iii)
; (iii) 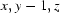 ; (iv)
; (iv) 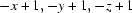 .
.
Acknowledgments
Financial support of this work by the Ministry of Education, Youth and Sports of the Czech Republic (MSM6198959218) and the Grant Agency of the Czech Republic (GAČR 203/08/P436) is gratefully acknowledged.
supplementary crystallographic information
Comment
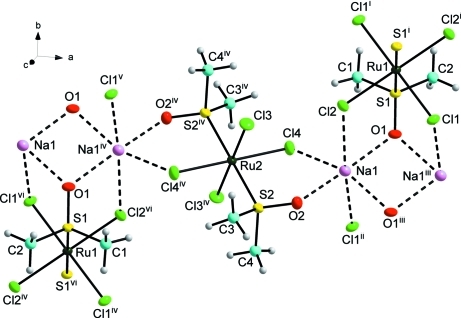
The title complex, (I), was prepared as a part of our study of simple ruthenium(III) complexes of the composition M[RuCl4(DMSO)2] with the different types of inorganic and organic cations [M]. In the crystal structure of (I), the centrosymmetric [RuCl2(DMSO)Na(DMSO)Cl2Ru] units are present; each ruthenium atom lies on a crystallographic centre of inversion whereas all other atoms lie in general positions. Each of the RuIII atoms is hexacoordinated by two sulphur atoms from two DMSO molecules in apical positions and four chlorido ligands in an equatorial plane (Fig. 1). Each ruthenium atom exists within an octahedral trans-RuCl4S2 donor set.
Both octahedra exhibit similar Ru—S bond lengths [Ru1—S1 = 2.3350 (5) and Ru2—S2= 2.3373 (6) Å] in contrast to the Ru—Cl bond lengths, which exhibit similar values in the Ru1 octahedron [2.3551 (5) and 2.3509 (5) Å] but differ significantly in the Ru2 octahedron [2.3353 (6) and 2.3663 (6) Å]. The Ru—Cl bond lengths within the [RuCl2(DMSO)Na(DMSO)Cl2Ru] unit do not differ markedly from those observed in Na[trans-RuCl4(DMSO)(pyr)].DMSO, where pyr = 1,4-pyrazine, [Ru—Cl = (2.3395 (8) - 2.3754 (7) Å] (Anderson et al., 2007) and in Na2[{trans-RuCl4(DMSO)}2(µ-pyrimidine)], where the Ru—Cl bond lengths are in the range of 2.338 (2) - 2.361 (2) Å (Iengo et al., 1999), while the Ru—S distances in (I) are slightly longer than those observed in the above-mentioned complexes (Ru—S are in the range of 2.281 (2) - 2.3027 (7) Å).
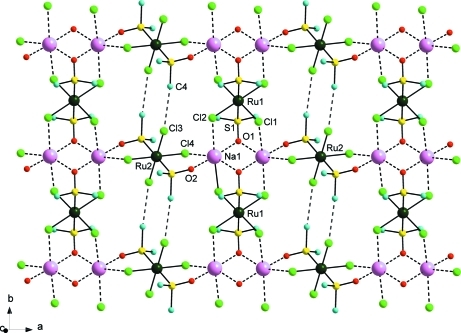
The Na cation has six atoms in its closest octahedrally coordinated environment with the resulting Cl3O3 donor set and it is surrounded by six donor interactions, represented by dashed lines (Figs. 1 and 2). The distance between the two nearest Na atoms equals 3.4968 (19) Å. The distances of the Na—O and Na—Cl donor interactions vary from 2.2974 (18) to 2.4155 (18) Å, and from 2.7773 (11) to 2.9374 (10) Å, respectively. Similar values of the Na—O distances [2.272 (4) - 2.418 (4) Å] were found in Na[trans-RuCl4(DMSO)(NH3)].DMSO (Alessio et al., 1993) and similar Na—Cl distances [2.776 (2) - 2.922 (2) Å] were observed in Na[trans-RuCl4(DMSO)(pyr)].DMSO (Anderson et al., 2007).
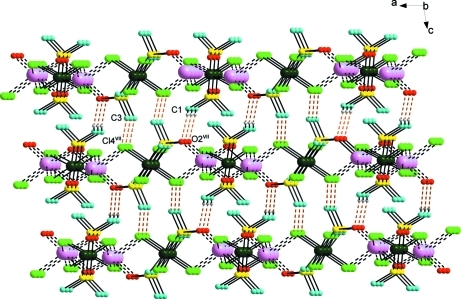
The trans-[Ru(DMSO)2Cl4] complex anion is connected to four sodium cations, two DMSO molecules and four chlorido ligands into infinite two dimensional array in the ab plane (Fig. 2). The crystal strucure of (I) is furter stabilized by non-bonding interactions of the C···Cl type [C4···Cl3ii is 3.284 (2) Å (ii = x, y+1, z)], which connect two Ru2 anions in the b direction (Fig. 2). The planes are cross connected, in the c direction, by the weak C···Cl [C3—Cl4vii is 3.903 (3) Å] and C—H···O [C1···O2vii is 3.376 (3) Å (vii = x, 1/2-y, -1/2+z)] type interactions (Fig. 3).
Experimental
The title complex was prepared by a slightly modified literature procedure (Alessio et al., 1991). The salt [H+(DMSO)2][trans-Ru(DMSO)2Cl4] (1 mmol) was dissolved in a mixture of ethanol (20 ml) and distilled water (0.3 ml). The orange solution was filtered, and then, an aqueous solution (0.3 ml) of NaCl (0.07 g, 2.8 mmol) was added during stirring. The yellow precipitate which formed after several minutes was filtered off, washed with cold acetone, and dried in air (yield 50%). Single crystals were obtained from the filtrate by slow evaporation after several days.
Refinement
All H-atoms were located in difference maps and refined using a riding model, with C–H distances of 0.98 Å, and with Uiso(H) = 1.5Ueq(C).
Figures
Fig. 1.
Part of the crystal structure of the title compound (I). The non-H atoms are drawn as 70% probability displacement ellipsoids. Symmetry codes: (i) -x+1, -y+1, -z+1; (ii) x, y+1, z; (iii) -x+1, -y, -z+1; (iv) -x, -y, 1-z; (v) x, y-1, z; (vi) -1+x, -1+y, z.
Fig. 2.
Part of the crystal structure of (I), showing the formation of the two dimensional array (a view along the c axis). Dashed lines represent non-covalent interactions of the C···Cl, Na···O and Na···Cl types. The H-atoms have been omitted for clarity. Symmetry code: (ii) x, 1+y, z.
Fig. 3.
Part of the crystal structure of (I), showing the formation of the two dimensional array (a view along the b axis). The H-atoms have been omitted for clarity. Dashed lines represent interactions of the C···Cl and C—H···O types. Symmetry code: (vii) x, 1/2-y, -1/2+z.
Crystal data
| [NaRuCl4(C2H6OS)2] | F(000) = 828 |
| Mr = 422.12 | Dx = 2.140 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 10079 reflections |
| a = 11.9042 (3) Å | θ = 2.9–32.0° |
| b = 8.0692 (2) Å | µ = 2.34 mm−1 |
| c = 13.7873 (3) Å | T = 120 K |
| β = 98.470 (2)° | Prism, orange |
| V = 1309.93 (5) Å3 | 0.20 × 0.20 × 0.15 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Xcalibur2 CCD diffractometer | 2300 independent reflections |
| Radiation source: fine-focus sealed tube | 2105 reflections with I > 2σ(I) |
| Enhance (Mo) X-ray Source | Rint = 0.021 |
| Detector resolution: 8.3611 pixels mm-1 | θmax = 25.0°, θmin = 2.9° |
| rotation method, ω–scan | h = −14→12 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007) | k = −9→9 |
| Tmin = 0.652, Tmax = 0.721 | l = −14→16 |
| 10197 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.018 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.050 | H-atom parameters constrained |
| S = 1.15 | w = 1/[σ2(Fo2) + (0.0269P)2 + 0.8654P] where P = (Fo2 + 2Fc2)/3 |
| 2300 reflections | (Δ/σ)max = 0.001 |
| 134 parameters | Δρmax = 0.71 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ru1 | 0.5000 | 0.5000 | 0.5000 | 0.00777 (8) | |
| S1 | 0.45936 (5) | 0.30580 (7) | 0.37416 (4) | 0.00948 (13) | |
| Cl1 | 0.35603 (5) | 0.67940 (7) | 0.42755 (4) | 0.01284 (13) | |
| Na1 | 0.35677 (8) | 0.00303 (10) | 0.51520 (7) | 0.0144 (2) | |
| O1 | 0.47177 (13) | 0.12823 (19) | 0.40510 (11) | 0.0125 (3) | |
| C1 | 0.31978 (19) | 0.3274 (3) | 0.30958 (17) | 0.0148 (5) | |
| H1A | 0.3050 | 0.2387 | 0.2608 | 0.022* | |
| H1B | 0.3125 | 0.4351 | 0.2765 | 0.022* | |
| H1C | 0.2648 | 0.3202 | 0.3558 | 0.022* | |
| Ru2 | 0.0000 | 0.0000 | 0.5000 | 0.00862 (9) | |
| S2 | 0.12479 (5) | −0.14419 (7) | 0.61717 (4) | 0.01206 (13) | |
| Cl2 | 0.63676 (5) | 0.64639 (7) | 0.42686 (4) | 0.01285 (13) | |
| O2 | 0.24699 (13) | −0.0998 (2) | 0.62514 (11) | 0.0147 (4) | |
| C2 | 0.5426 (2) | 0.3346 (3) | 0.27921 (16) | 0.0145 (5) | |
| H2A | 0.5163 | 0.2593 | 0.2249 | 0.022* | |
| H2B | 0.6224 | 0.3112 | 0.3042 | 0.022* | |
| H2C | 0.5352 | 0.4494 | 0.2559 | 0.022* | |
| Cl3 | 0.07120 (5) | 0.23663 (7) | 0.58529 (4) | 0.01694 (14) | |
| C3 | 0.0887 (2) | −0.1226 (4) | 0.73698 (18) | 0.0224 (6) | |
| H3A | 0.1455 | −0.1794 | 0.7841 | 0.034* | |
| H3B | 0.0138 | −0.1718 | 0.7390 | 0.034* | |
| H3C | 0.0868 | −0.0048 | 0.7539 | 0.034* | |
| Cl4 | −0.14497 (5) | −0.01787 (7) | 0.59997 (4) | 0.01570 (14) | |
| C4 | 0.1151 (2) | −0.3619 (3) | 0.6015 (2) | 0.0262 (6) | |
| H4A | 0.1588 | −0.4168 | 0.6583 | 0.039* | |
| H4B | 0.1458 | −0.3934 | 0.5420 | 0.039* | |
| H4C | 0.0354 | −0.3960 | 0.5956 | 0.039* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ru1 | 0.00907 (15) | 0.00689 (15) | 0.00724 (14) | 0.00048 (9) | 0.00081 (11) | 0.00072 (9) |
| S1 | 0.0118 (3) | 0.0075 (3) | 0.0088 (3) | 0.0004 (2) | 0.0006 (2) | 0.0000 (2) |
| Cl1 | 0.0137 (3) | 0.0105 (3) | 0.0133 (3) | 0.0032 (2) | −0.0017 (2) | 0.0002 (2) |
| Na1 | 0.0131 (5) | 0.0128 (5) | 0.0173 (5) | −0.0001 (3) | 0.0023 (4) | 0.0027 (4) |
| O1 | 0.0165 (9) | 0.0073 (8) | 0.0136 (8) | 0.0005 (6) | 0.0015 (7) | 0.0005 (6) |
| C1 | 0.0134 (12) | 0.0148 (12) | 0.0148 (12) | 0.0005 (10) | −0.0027 (10) | −0.0016 (10) |
| Ru2 | 0.00868 (15) | 0.00670 (15) | 0.01060 (15) | −0.00093 (9) | 0.00177 (11) | −0.00048 (9) |
| S2 | 0.0113 (3) | 0.0106 (3) | 0.0142 (3) | −0.0004 (2) | 0.0014 (2) | 0.0025 (2) |
| Cl2 | 0.0140 (3) | 0.0126 (3) | 0.0126 (3) | −0.0021 (2) | 0.0042 (2) | 0.0007 (2) |
| O2 | 0.0108 (8) | 0.0183 (9) | 0.0148 (8) | 0.0001 (7) | 0.0013 (7) | 0.0030 (7) |
| C2 | 0.0195 (13) | 0.0135 (12) | 0.0110 (11) | −0.0006 (10) | 0.0042 (10) | −0.0009 (9) |
| Cl3 | 0.0202 (3) | 0.0107 (3) | 0.0194 (3) | −0.0040 (2) | 0.0013 (3) | −0.0048 (2) |
| C3 | 0.0176 (13) | 0.0349 (16) | 0.0147 (12) | 0.0010 (11) | 0.0022 (11) | 0.0066 (11) |
| Cl4 | 0.0116 (3) | 0.0220 (3) | 0.0142 (3) | −0.0003 (2) | 0.0040 (2) | 0.0019 (2) |
| C4 | 0.0267 (15) | 0.0113 (13) | 0.0383 (16) | 0.0002 (11) | −0.0032 (13) | 0.0039 (11) |
Geometric parameters (Å, °)
| Ru1—S1 | 2.3350 (5) | Ru2—Cl3iv | 2.3353 (6) |
| Ru1—S1i | 2.3351 (5) | Ru2—Cl3 | 2.3353 (6) |
| Ru1—Cl1 | 2.3509 (5) | Ru2—S2iv | 2.3373 (6) |
| Ru1—Cl1i | 2.3509 (5) | Ru2—S2 | 2.3373 (6) |
| Ru1—Cl2i | 2.3551 (5) | Ru2—Cl4iv | 2.3663 (6) |
| Ru1—Cl2 | 2.3551 (5) | Ru2—Cl4 | 2.3663 (6) |
| S1—O1 | 1.4962 (16) | S2—O2 | 1.4863 (16) |
| S1—C2 | 1.770 (2) | S2—C4 | 1.772 (3) |
| S1—C1 | 1.774 (2) | S2—C3 | 1.776 (2) |
| Cl1—Na1ii | 2.8769 (10) | Cl2—Na1i | 2.9374 (10) |
| Na1—O2 | 2.2974 (18) | C2—H2A | 0.9800 |
| Na1—O1 | 2.4105 (17) | C2—H2B | 0.9800 |
| Na1—O1iii | 2.4155 (18) | C2—H2C | 0.9800 |
| Na1—Cl4iv | 2.7773 (11) | C3—H3A | 0.9800 |
| Na1—Cl1v | 2.8769 (10) | C3—H3B | 0.9800 |
| Na1—Cl2i | 2.9374 (10) | C3—H3C | 0.9800 |
| Na1—Na1iii | 3.4968 (19) | Cl4—Na1iv | 2.7772 (11) |
| O1—Na1iii | 2.4156 (18) | C4—H4A | 0.9800 |
| C1—H1A | 0.9800 | C4—H4B | 0.9800 |
| C1—H1B | 0.9800 | C4—H4C | 0.9800 |
| C1—H1C | 0.9800 | ||
| S1—Ru1—S1i | 179.999 (1) | S1—C1—H1B | 109.5 |
| S1—Ru1—Cl1 | 92.250 (19) | H1A—C1—H1B | 109.5 |
| S1i—Ru1—Cl1 | 87.751 (19) | S1—C1—H1C | 109.5 |
| S1—Ru1—Cl1i | 87.751 (19) | H1A—C1—H1C | 109.5 |
| S1i—Ru1—Cl1i | 92.248 (19) | H1B—C1—H1C | 109.5 |
| Cl1—Ru1—Cl1i | 180.0 | Cl3iv—Ru2—Cl3 | 179.999 (1) |
| S1—Ru1—Cl2i | 84.313 (19) | Cl3iv—Ru2—S2iv | 84.97 (2) |
| S1i—Ru1—Cl2i | 95.687 (19) | Cl3—Ru2—S2iv | 95.03 (2) |
| Cl1—Ru1—Cl2i | 89.091 (19) | Cl3iv—Ru2—S2 | 95.03 (2) |
| Cl1i—Ru1—Cl2i | 90.909 (19) | Cl3—Ru2—S2 | 84.97 (2) |
| S1—Ru1—Cl2 | 95.686 (19) | S2iv—Ru2—S2 | 179.999 (1) |
| S1i—Ru1—Cl2 | 84.314 (19) | Cl3iv—Ru2—Cl4iv | 89.90 (2) |
| Cl1—Ru1—Cl2 | 90.909 (19) | Cl3—Ru2—Cl4iv | 90.10 (2) |
| Cl1i—Ru1—Cl2 | 89.091 (19) | S2iv—Ru2—Cl4iv | 90.63 (2) |
| Cl2i—Ru1—Cl2 | 180.0 | S2—Ru2—Cl4iv | 89.37 (2) |
| O1—S1—C2 | 107.12 (10) | Cl3iv—Ru2—Cl4 | 90.10 (2) |
| O1—S1—C1 | 106.43 (10) | Cl3—Ru2—Cl4 | 89.90 (2) |
| C2—S1—C1 | 101.59 (11) | S2iv—Ru2—Cl4 | 89.37 (2) |
| O1—S1—Ru1 | 115.46 (7) | S2—Ru2—Cl4 | 90.63 (2) |
| C2—S1—Ru1 | 112.59 (8) | Cl4iv—Ru2—Cl4 | 180.0 |
| C1—S1—Ru1 | 112.54 (8) | O2—S2—C4 | 107.04 (12) |
| Ru1—Cl1—Na1ii | 115.06 (3) | O2—S2—C3 | 106.03 (11) |
| O2—Na1—O1 | 176.27 (7) | C4—S2—C3 | 100.97 (14) |
| O2—Na1—O1iii | 93.79 (6) | O2—S2—Ru2 | 116.54 (7) |
| O1—Na1—O1iii | 87.14 (6) | C4—S2—Ru2 | 112.72 (10) |
| O2—Na1—Cl4iv | 80.56 (5) | C3—S2—Ru2 | 112.19 (9) |
| O1—Na1—Cl4iv | 100.01 (5) | Ru1—Cl2—Na1i | 111.02 (3) |
| O1iii—Na1—Cl4iv | 156.08 (5) | S2—O2—Na1 | 133.14 (10) |
| O2—Na1—Cl1v | 88.98 (5) | S1—C2—H2A | 109.5 |
| O1—Na1—Cl1v | 94.75 (5) | S1—C2—H2B | 109.5 |
| O1iii—Na1—Cl1v | 75.12 (4) | H2A—C2—H2B | 109.5 |
| Cl4iv—Na1—Cl1v | 81.52 (3) | S1—C2—H2C | 109.5 |
| O2—Na1—Cl2i | 99.39 (5) | H2A—C2—H2C | 109.5 |
| O1—Na1—Cl2i | 76.89 (4) | H2B—C2—H2C | 109.5 |
| O1iii—Na1—Cl2i | 108.18 (5) | S2—C3—H3A | 109.5 |
| Cl4iv—Na1—Cl2i | 95.69 (3) | S2—C3—H3B | 109.5 |
| Cl1v—Na1—Cl2i | 170.66 (4) | H3A—C3—H3B | 109.5 |
| O2—Na1—Na1iii | 137.18 (6) | S2—C3—H3C | 109.5 |
| O1—Na1—Na1iii | 43.63 (4) | H3A—C3—H3C | 109.5 |
| O1iii—Na1—Na1iii | 43.51 (4) | H3B—C3—H3C | 109.5 |
| Cl4iv—Na1—Na1iii | 138.71 (5) | Ru2—Cl4—Na1iv | 110.05 (3) |
| Cl1v—Na1—Na1iii | 83.09 (3) | S2—C4—H4A | 109.5 |
| Cl2i—Na1—Na1iii | 93.39 (4) | S2—C4—H4B | 109.5 |
| S1—O1—Na1 | 122.65 (9) | H4A—C4—H4B | 109.5 |
| S1—O1—Na1iii | 126.22 (9) | S2—C4—H4C | 109.5 |
| Na1—O1—Na1iii | 92.86 (6) | H4A—C4—H4C | 109.5 |
| S1—C1—H1A | 109.5 | H4B—C4—H4C | 109.5 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y+1, z; (iii) −x+1, −y, −z+1; (iv) −x, −y, −z+1; (v) x, y−1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2632).
References
- Alessio, E., Balducci, G., Calligaris, M., Costa, G., Attia, G. M. & Mestroni, G. (1993). Inorg. Chim. Acta, 30, 609–618.
- Alessio, E., Balducci, G., Lutman, A., Mestroni, G., Calligaris, M. & Attia, G. M. (1991). Inorg. Chim. Acta, 203, 205–217.
- Anderson, C. M., Herman, A. & Rochon, F. D. (2007). Polyhedron, 26, 3661–3668.
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Iengo, E., Mestroni, G., Geremia, S., Calligaris, M. & Alessio, E. (1999). J. Chem. Soc. Dalton Trans. pp. 3361–3371.
- Oxford Diffraction (2007). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Abingdon, England.
- Piggot, P. M. T., Hall, L. A., White, A. J. P. & Williams, D. J. (2004). Inorg. Chim. Acta, 357, 250–258.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810007063/tk2632sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810007063/tk2632Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report