Abstract
Signature databases are vital tools for identifying distant relationships in novel sequences and hence for inferring protein function. InterPro is an integrated documentation resource for protein families, domains and functional sites, which amalgamates the efforts of the PROSITE, PRINTS, Pfam and ProDom database projects. Each InterPro entry includes a functional description, annotation, literature references and links back to the relevant member database(s). Release 2.0 of InterPro (October 2000) contains over 3000 entries, representing families, domains, repeats and sites of post-translational modification encoded by a total of 6804 different regular expressions, profiles, fingerprints and Hidden Markov Models. Each InterPro entry lists all the matches against SWISS-PROT and TrEMBL (more than 1 000 000 hits from 462 500 proteins in SWISS-PROT and TrEMBL). The database is accessible for text- and sequence-based searches at http://www.ebi.ac.uk/interpro/. Questions can be emailed to interhelp@ebi.ac.uk.
INTRODUCTION
Databases with signatures diagnostic for protein families, domains or functional sites are important tools for the computational functional classification of newly determined sequences that lack biochemical characterisation. During the last decade, several signature recognition and sequence clustering methods have evolved to address different sequence analysis problems, resulting in rather different and, for the most part, independent databases. Currently, the most commonly used signature and sequence cluster databases include PROSITE (1); Pfam (2); PRINTS (3); ProDom (4); and Blocks (5). Diagnostically, these resources have different areas of optimum application owing to the different strengths and weaknesses of their underlying analysis methods.
In terms of family coverage, the signature databases are similar in size but differ in content. While all of the resources share a common interest in protein sequence classification, the focus of each database is different. Pfam, for example, focuses on divergent domains, PROSITE on functional sites and PRINTS focuses on families, specialising in hierarchical definitions from super-family down to sub-family levels in order to describe specific functions. A number of sequence cluster databases, for example ProDom, are also commonly used in sequence analysis to facilitate domain identification. Unlike signature databases, the clustered resources are derived automatically from sequence databases, using different clustering algorithms. Databases like Blocks provide ungapped multiple alignments for protein families.
With the rapid release of raw data from genome sequencing projects, there is a strong dependence on automatic methods for assigning functions to unknown sequences. For this sequence characterisation, we need more reliable, concerted methods for identifying protein family traits and for inheriting functional annotation. InterPro was developed to rationalise this process by creating a single coherent resource for diagnosis and documentation of protein families. This new resource provides an integrated view of a number of commonly used signature databases and provides an intuitive interface for text- and sequence-based searches.
INTEGRATION METHODS
Flat-files submitted by each of the member databases, PRINTS, PROSITE, Pfam and ProDom, were systematically merged and dismantled. Overlapping domains, signatures or profiles describing common domains or protein families were merged into a single InterPro entry with a unique accession number (which takes the form IPRxxxxxx, where x is a digit), while those containing no counterpart in other member databases were assigned their own unique accession numbers. This process was complicated by the relationships that can exist, both between entries in the same database and between entries in different databases. Different types of hierarchical family relationships were evident, leading us to recognise ‘sub-types’ and ‘sub-strings’. A sub-string means that a motif or motifs are contained within a region of sequence encoded by a wider pattern (e.g. a PROSITE pattern is typically contained within a PRINTS fingerprint; or a fingerprint might be contained within a Pfam domain). A sub-type means that one or more motifs are specific for a sub-set of sequences captured by another more general pattern and these are described as ‘parent–child’ relationships. Signatures with sub-string relationships have the same IPR numbers, while sub-type parent–child relationships warrant their own IPRs. The domain structure of multidomain proteins is described in a ‘contains/found in’ relationship, where a set of family signatures can contain InterPro entries describing specific domains, but they are not related in the protein family sense. These relationships are demonstrated in Figure 1.
Figure 1.
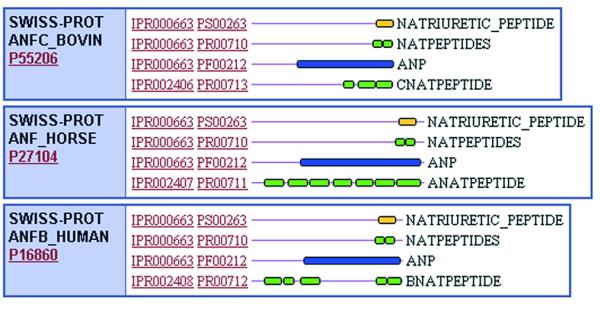
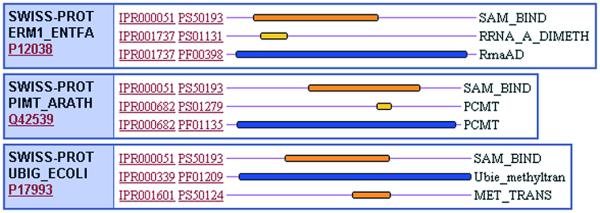
Demonstration of relationships existing between InterPro entries. (Top) Parent–child relationship. This graphical view of three proteins shows IPR000663, which contains signatures describing the Natriuretic peptide family. Each protein has an additional InterPro entry associated with it, containing a fingerprint for more specific classes of Natriuretic peptide. These InterPro entries, IPR002406, IPR002407 and IPR002408 are the children or sub-families of IPR000663. (Bottom) Contains-found in relationship. In these three proteins, IPR000051, the SAM binding motif is a domain found in several different protein families, including IPR001737 (ribosomal RNA adenine dimethylase), IPR000682 (protein-l-isoaspartate(d-aspartate) O-methyltransferase) and IPR000339, a family of ubiqunone methyltransferases. They are not sub-families of the SAM binding domain.
CONTENTS OF CURRENT RELEASE
Release 2.0 of InterPro was built from Pfam 5.5 (2479 domains), PRINTS 27 (1356 fingerprints), ProDom 2000.1 (1309 domains), PROSITE 16.25 (1424 patterns and profiles) and 236 preliminary profiles. The release contains 3203 entries with 1 315 676 hits in SWISS-PROT and TrEMBL (6). Of these hits, 1 244 893 are considered to be true, 9303 false positive, 4524 false negative, 2885 are partial hits and 54 071 have the status unknown. The SWISS-PROT and TrEMBL match lists are provided by the member databases. An exception here concerns PROSITE pattern hits against TrEMBL, which undergo a different procedure. These are not provided by PROSITE and must therefore be derived by the TrEMBL group. All TrEMBL entries are scanned for PROSITE patterns. If a match is found, its significance is checked by means of a set of secondary patterns computed with the eMOTIF algorithm (7). For each family in PROSITE, the true members are aligned and fed into eMOTIF, which calculates a near optimal set of regular expressions, based on statistical rather than biological evidence. A stringency of 10–9 is used, so that each eMOTIF pattern is expected to produce a random or false positive hit in 10–9 matches. All pattern hits confirmed by eMOTIF are considered true; all others are flagged as unknown.
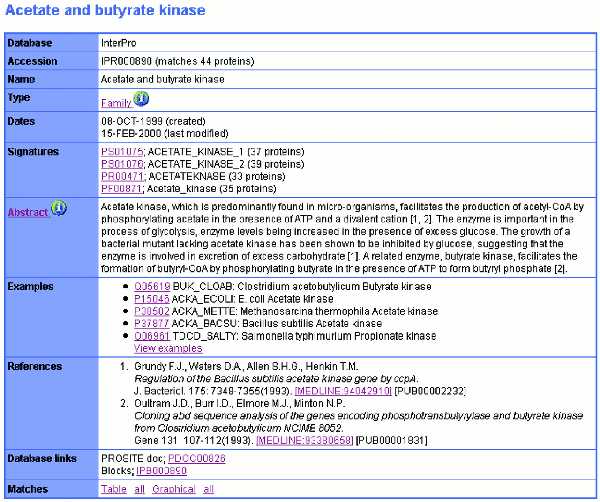
Individual InterPro entries contain a description of the protein family, domain, repeat or post-translational modification (e.g. N-glycosylation site); a list of member database signatures, Hidden Markov Models (HMMs), profiles or fingerprints associated with the entry; an abstract derived from merged annotation from the member databases; examples of representative sequences; literature references used to create the abstract; and links to tabular or graphical views of the matches to SWISS-PROT and TrEMBL. An example is shown in Figure 2.
Figure 2.
An example of an InterPro entry. This is IPR000890, an entry containing signatures describing the acetate and butyrate kinase protein family. The ‘i’ information buttons have links to help files describing, for example, the ‘Family’ concept.
DATABASE FORMAT, ACCESS AND DISTRIBUTION
To facilitate in-house maintenance, InterPro is managed within a relational database system. However, the InterPro database is also released in two ASCII (text) flat-files in XML (eXtended Markup Language) format, one containing the core InterPro entries and the other containing the protein matches. These come together with a corresponding DTD (Document Type Definition) file, to allow users to keep local InterPro copies on their machines. The InterPro flat-file may be retrieved from the EBI anonymous ftp server (ftp://ftp.ebi.ac.uk/pub/databases/interpro).
InterPro is accessible for interactive use via the EBI Web server (http://www.ebi.ac.uk/interpro), which can also be reached via each of the member databases. The Web interface allows text-based and sequence-based searches using a sequence retrieval system (SRS) (8). The sequence-based searches are done using InterProScan, which combines the search methods from the member databases. The results display matches to the parent databases and the corresponding InterPro entries, providing the positions of the signatures within the sequence and a graphical view of the matches. Detailed results of matches to the individual database search methods are provided via hyperlinks to each of the parent databases. A mail server is available for sequence searches at interproscan@ebi.ac.uk. Documentation on using the mail server can be obtained by emailing the address with the word ‘help’ in the body of the text.
APPLICATIONS OF INTERPRO
InterPro is an international initiative that was conceived in an attempt to streamline the efforts of the signature database providers. By uniting these databases, we capitalise on their individual strengths, producing a single entity that is far greater than the sum of its parts. A primary application of InterPro’s family, domain and functional site definitions will be in annotation and functional classification of uncharacterised sequences. The EBI is using InterPro for enhancing the automated annotation of TrEMBL (9). This is more efficient and reliable than using each of the signature databases separately, because InterPro provides internal consistency checks and deeper coverage. InterPro has also proven its usefulness for whole proteome analysis in the comparative genome analysis of Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae (10).
Another major use of InterPro will be in identifying those families and domains for which the existing discriminators are not optimal and could hence be usefully supplemented with an alternative pattern (e.g. where a regular expression identifies large numbers of false matches it could be useful to develop an HMM or where an HMM covers a vast super-family it could be beneficial to develop discrete family fingerprints, and so on). Alternatively, InterPro is likely to highlight key areas where none of the databases has yet made a contribution and hence where the development of a specific pattern might be useful. For example, sequence groups from ProDom are being analysed using the Pratt pattern discovery tool (11,12) to reveal clusters that can form InterPro families and to create regular expression discriminators. This united approach should thus help us to improve both the utility and the coverage of signature databases, pinpointing weaknesses and allowing us to remedy them efficiently.
As it evolves, InterPro will streamline the analysis of newly determined sequences for the individual user and will make a significant contribution in the demanding task of automatic classification of predicted proteins from genome sequencing projects.
FUTURE DIRECTIONS
The InterPro project began by first integrating the databases that provide annotation (Pfam, PRINTS and PROSITE). Various factors rendered a step-wise approach to the development of InterPro desirable. First, the scale of the task of amalgamating the first three databases was immense. The rational merging of apparently equivalent database entries that in fact simultaneously define a specific family, domains within that family or even repeats within those domains, presented an enormous challenge. A second important consideration was that while Pfam, PRINTS and PROSITE are true pattern databases, ProDom is based solely on automatic clustering of sequences by similarity (i.e. discriminators are not derived). Resulting clusters need not have precise biological correlations and some family designations have changed between database versions. The initial integration of ProDom has therefore been limited to well-defined protein families and those entries with corresponding overlaps in the other member databases. The next goal is the further integration of ProDom entries.
In addition, the Blocks database is now using InterPro to replace their old Blocks from PROSITE (J.Henikoff, personal communication). As the current and subsequent Blocks releases will be based on families already in InterPro, the process of cross-referencing between Blocks and InterPro was relatively straightforward and was done for the current InterPro release. Once the founder members of the InterPro consortium have been assimilated into the unified resource, other pattern databases will also be included. First, scheduled for Release 3, will be the SMART resource (13). Ultimately, we hope to include many other protein family databases to give a more comprehensive view of the resources available.
Acknowledgments
ACKNOWLEDGEMENTS
The InterPro project is supported by grant number BIO4-CT98-0052 of the European Commission. T.K.A. is a Royal Society University Research Fellow.
References
- 1.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L.L. (2000) The Pfam Protein Families Database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attwood T.K., Croning,M.D.R., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J.N. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nevill-Manning C.G., Wu,T.D. and Brutlag,D.L. (1998) Highly specific protein sequence motifs for genome analysis. Proc. Natl Acad. Sci. USA, 95, 5865–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etzold,T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol ., 266, 114–128. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann W., Möller,S., Gateau,A. and Apweiler R. (1999) A novel method for automatic functional annotation of proteins. Bioinformatics, 15, 228–233. [DOI] [PubMed] [Google Scholar]
- 10.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonassen I., Collins,J.F. and Higgins,D. (1995) Finding flexible patterns in unaligned protein sequences. Protein Sci., 4, 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonassen I. (1997) Efficient discovery of conserved patterns using a pattern graph. Comput. Appl. Biosci., 13, 509–522. [DOI] [PubMed] [Google Scholar]
- 13.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]