Abstract
The transcriptional activity of p53 is regulated by a cascade of posttranslational modifications. Although acetylation of p53 by CREB-binding protein (CBP)/p300 is known to be indispensable for p53 activation, the role of phosphorylation, and in particular multisite phosphorylation, in activation of CBP/p300-dependent p53 transcriptional pathways remains unclear. We investigated the role of single site and multiple site phosphorylation of the p53 transactivation domain in mediating its interaction with CBP and with the ubiquitin ligase HDM2. Phosphorylation at Thr18 functions as an on/off switch to regulate binding to the N-terminal domain of HDM2. In contrast, binding to CBP is modulated by the extent of p53 phosphorylation; addition of successive phosphoryl groups enhances the affinity for the TAZ1, TAZ2, and KIX domains of CBP in an additive manner. Activation of p53-dependent transcriptional pathways requires that p53 compete with numerous cellular transcription factors for binding to limiting amounts of CBP/p300. Multisite phosphorylation represents a mechanism for a graded p53 response, with each successive phosphorylation event resulting in increasingly efficient recruitment of CBP/p300 to p53-regulated transcriptional programs, in the face of competition from cellular transcription factors. Multisite phosphorylation thus acts as a rheostat to enhance binding to CBP/p300 and provides a plausible mechanistic explanation for the gradually increasing p53 response observed following prolonged or severe genotoxic stress.
Keywords: competitive binding, protein–protein interaction, transcriptional coactivator, tumor suppressor
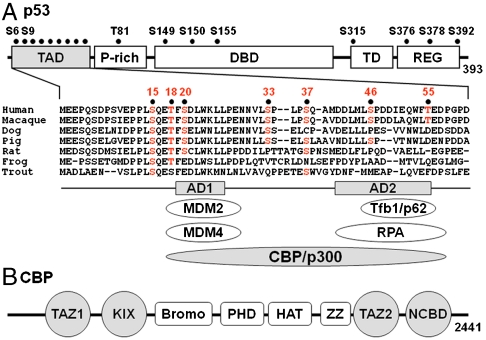
The transcriptional activity of the tumor suppressor p53 is triggered in response to genotoxic stress caused by DNA damage. p53 is maintained at low levels in unstressed cells through continuous proteasomal degradation mediated by the E3 ubiquitin ligase HDM2 (the human homolog of mouse double minute 2, MDM2). DNA damage initiates a cascade of phosphorylation and acetylation events that result in stabilization and accumulation of p53 and lead to arrest of cell growth and apoptosis (1–5). p53 is tightly regulated through coordinated interactions with HDM2 and the general transcriptional coactivators c-AMP response element binding (CREB) binding protein (CBP) and p300. The interactions are mediated by the N-terminal transcriptional activation domain (TAD) of p53, which is intrinsically disordered (6, 7) and utilizes two subdomains (AD1 and AD2) to bind the N-terminal domain of HDM2 and the TAZ1, TAZ2, KIX, and nuclear receptor coactivator binding (NCBD) domains of CBP/p300 (Fig. 1) (8, 9). In unstressed cells, p53 forms a ternary complex with CBP/p300 and HDM2 that promotes polyubiquitination and degradation of p53 (9, 10). Phosphorylation of p53 at Thr18 following DNA damage impairs binding to the N-terminal region of HDM2 while enhancing binding to the CBP TAZ1, TAZ2, and KIX domains (9, 11). Although single-site phosphorylation of p53 at Thr18 causes only twofold increase in affinity to CBP TAZ1, triple phosphorylation at Ser15, Thr18, and Ser20 results in more than 10-fold enhancement in binding affinity (9).
Fig. 1.
Domain organization of p53 and CBP/p300. (A) Schematic of p53 showing TAD (N-terminal transactivation domain), P-rich (proline-rich), DBD (DNA-binding domain), TD (tetramerization domain), and REG (C-terminal regulatory) domains. The sequence alignment of p53 TAD is shown for a few species, and known sites of phosphorylation are indicated by black dots. The AD1 and AD2 motifs are indicated, and proteins that interact directly with them are shown below the sequence alignment. (B) Domains of CBP/p300. The TAZ1 (residues 340–439), KIX (586–672), TAZ2 (1,764–1,855), and NCBD (2,059–2,117) domains interact with p53 TAD and are highlighted in gray.
Although it is clear that p53 is activated by a complex cascade of phosphorylation and other posttranslational modifications initiated by genotoxic stress, the functional roles played by individual phosphorylation sites and by multisite phosphorylation events remain poorly understood (1, 2, 4, 12). Nine of the 17 known phosphorylation sites in human p53 are located in the TAD (Ser6, Ser9, Ser15, Thr18, Ser20, Ser33, Ser37, Ser46, and Thr55) (Fig. 1); simultaneous phosphorylation at two to four sites in the N-terminal region (residues 1–24) of the p53 TAD has been observed in cell extracts (13). Single-site phosphorylation increases the affinity of p53 for binding to CBP/p300 in vitro (11, 14–16) or in vivo (17). Phosphorylation of Thr18, which requires prior phosphorylation of Ser15 by the ATM/ATR kinases or DNA-PK (18), impairs binding to HDM2 in vitro and may help to stabilize p53 (19, 20). Studies of knock-in mice, in which Ser18 or Ser23 (equivalent to Ser15 and Ser20 in human p53) are substituted by alanine, show that loss of these phosphorylation sites partially compromises the ability of p53 to induce apoptosis in response to ionizing radiation (21). Overall, however, the phenotypical changes resulting from targeted single-site mutations of individual serines and threonines are modest, raising questions about the role of single-site phosphorylation events in vivo (4, 12). Simultaneous mutation of Ser18 and Ser23 to alanine leads to more severe defects in mice, suggesting that two-site or multisite phosphorylation has synergistic effects in activating the p53 response (22). When simultaneously phosphorylated, Thr18 and Ser20 in human p53 also appear to function synergistically to enhance the p53 response (23, 24).
To obtain insights into the role of combinatorial phosphorylation in modulating interactions of p53 with HDM2 and CBP/p300, we systematically assessed the effect of single-, double-, and triple-site phosphorylation of the p53 TAD upon binding to the N-terminal domain of HDM2 and to the TAZ1, TAZ2, KIX, and NCBD domains of CBP. Our data complement and extend recent work by Teufel et al. (11) on the effect of phosphorylation upon binding to p300, showing conclusively that multisite phosphorylation enhances binding of p53 to CBP/p300 through a graded response that is proportional to the number of phosphoryl groups. Multisite phosphorylation functions as a rheostat that enhances the ability of p53 to compete with cellular transcription factors for binding to limiting amounts of CBP/p300 and may control the nature and extent of the p53 response to prolonged genotoxic stress.
Results and Discussion
Effect of p53 TAD Phosphorylation on Binding to CBP and HDM2.
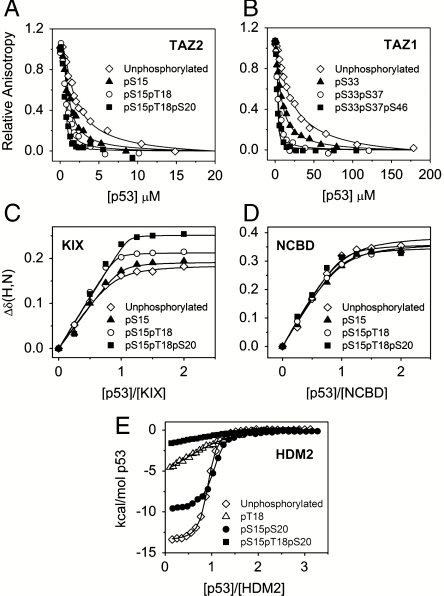
Single-, double-, and triple-phosphorylated p53 TAD peptides were generated and interactions with the CBP TAZ1 and TAZ2 domains were monitored by fluorescence anisotropy. Interactions with the KIX and NCBD domains were monitored by NMR titrations, and binding to the N-terminal domain of HDM2 was studied by isothermal titration calorimetry (ITC). Representative titration curves are shown in Fig. 2, and the measured dissociation constants (Kd) are given in Table 1. Consistent with previous reports (8, 9, 11), the unphosphorylated p53 TAD binds with highest affinity to the TAZ2 domain of CBP/p300 and more weakly to the TAZ1, KIX, and NCBD domains. Single-site phosphorylation at Ser15, Thr18, Ser20, Ser33, Ser37, and Ser46 increases the affinity for TAZ1 and TAZ2 by 2- to 4-fold and for KIX by 2- to 8-fold, but has only a small effect on binding to the NCBD (Table 1). Thr55 phosphorylation, which occurs constitutively in cells (25), has no effect on TAZ2 binding but enhances binding to TAZ1 and KIX by 2- and 4-fold, respectively. Two-site phosphorylation of the p53 TAD (pS15pT18, pS15pS20, pT18pS20, pS33pS37, pS33pS46, and pS37pS46) enhances the affinity for the TAZ1 and TAZ2 domains by an average of 11- and 12-fold, respectively, whereas triple phosphorylation at Ser15, Thr18, and Ser20 or Ser33, Ser37, and Ser46 further increases the affinity (an average of 20-fold and 25-fold for TAZ1 and TAZ2, respectively). Because binding to TAZ2 is so tight (Kd < 20 nM), the gain in affinity resulting from double and triple phosphorylation was not observed in previously reported NMR titration experiments (9). Double phosphorylation at S15T18, S15S20, and T18S20 increases the affinity for KIX by 12-fold on average, with a further increase upon simultaneous phosphorylation at all three sites. However, TAD peptides phosphorylated at S33 and S46 or at S37 and S46 bind no more tightly than their singly phosphorylated counterparts. Note that, because KIX has two competing p53 binding sites of comparable affinity, the Kd values in Table 1 represent apparent dissociation constants, which are the harmonic mean of the Kds for each site (26).
Fig. 2.
Binding of unphosphorylated and phosphorylated p53 TAD to CBP domains and HDM2. (A) Representative fluorescence anisotropy titration curves for the competition of unlabeled p53 peptides (unphosphorylated p53 TAD, pS15, pS15pT18, pS15pT18pS20) with fluorescently labeled p53 TAD bound to TAZ2. (B) Representative fluorescence anisotropy titration curves for the competition of unlabeled p53 peptides (unphosphorylated p53, pS33, pS33pS37, pS33pS37pS46) with fluorescently labeled p53 TAD bound to TAZ1. (C) HSQC titration curves for Arg671 of KIX showing the weighted average of the 15N and 1H chemical shift changes, Δδ(N,H)av, as a function of concentration ratio for the titration of 15N-labeled KIX with unphosphorylated and phosphorylated p53 peptides (pS15, pS15pT18, pS15pT18pS20). (D) Titration curves for Met2098 in the NCBD showing the average of the 15N and 1H chemical shift changes [Δδ(N,H)av] as a function of concentration ratio for the titration of 15N-labeled NCBD with unphosphorylated and phosphorylated p53 peptides (pS15, pS15pT18, pS15pT18pS20). Δδ(N,H)av = [(ΔδHN)2 + (ΔδN/5)2]1/2. Values of Δδ(N,H)av are plotted as symbols, and the continuous lines show the curves fitted globally to a one-site binding model. (E) ITC data for binding of HDM2 to unphosphorylated and phosphorylated p53 peptides (pT18, pS15pS20, pS15pT18pS20).
Table 1.
Dissociation constants for the interactions of the phosphorylated p53 TAD peptides with CBP and HDM2 domains
| Fluorescence |
NMR |
ITC |
||||||||
| p53 peptides | CBP TAZ2 |
CBP TAZ1 |
CBP KIX |
CBP NCBD |
HDM2 |
|||||
| Kd, nM | F* | Kd, nM | F | Kd (app)†, nM | F (app) | Kd, nM | F | Kd, nM | F | |
| (13-61) | 20 ± 1.6‡ | — | 920 ± 100 | — | 9,400 ± 500 | — | 9,300 ± 1,000 | — | 230 ± 20 | — |
| (13-57)pS15 | 6.8 ± 1.6 | 2.9 | 600 ± 90 | 1.5 | 2,200 ± 200 | 4.3 | 8,600 ± 1,300 | 1.1 | 450 ± 40 | 0.51 |
| (13-57)pT18 | 6.5 ± 1.1 | 3.1 | 460 ± 50 | 2.0 | 2,800 ± 200 | 3.4 | 1,5700 ± 1,800 | 0.6 | 5,200 ± 1,300 | 0.04 |
| (13-57)pS20 | 5.3 ± 1.3 | 3.8 | 440 ± 60 | 2.1 | 1,200 ± 200 | 7.8 | 6,100 ± 1,200 | 1.5 | 350 ± 10 | 0.66 |
| (13-57)pS33 | 9.4 ± 1.3 | 2.1 | 350 ± 50 | 2.6 | 1,800 ± 300 | 5.2 | 5,200 ± 1,100 | 1.8 | 280 ± 10 | 0.82 |
| (13-57)pS37 | 14 ± 1.9 | 1.4 | 450 ± 60 | 2.0 | 2,500 ± 400 | 3.8 | 5,000 ± 900 | 1.9 | 300 ± 50 | 0.77 |
| (13-57)pS46 | 7.8 ± 1.0 | 2.6 | 210 ± 30 | 4.4 | 4,700 ± 600 | 2.0 | 4,000 ± 1,000 | 2.3 | 380 ± 30 | 0.61 |
| (13-57)pT55 | 22 ± 0.9 | 0.9 | 520 ± 90 | 1.8 | 1,900 ± 400 | 4.9 | 5,700 ± 1100 | 1.6 | 370 ± 20 | 0.62 |
| (13-57)pS15pT18 | 2.5 ± 0.9 | 8.0 | 200 ± 30 | 4.6 | 600 ± 90 | 16 | 5,100 ± 900 | 1.8 | 5,100 ± 800 | 0.05 |
| (13-57)pS15pS20 | 1.2 ± 0.7 | 16.7 | 140 ± 20 | 6.6 | 1,300 ± 100 | 7.2 | 3,400 ± 700 | 2.7 | 350 ± 10 | 0.66 |
| (13-57)pT18pS20 | 2.5 ± 1.1 | 8.0 | 110 ± 20 | 8.4 | 650 ± 140 | 15 | 3,600 ± 700 | 2.6 | 6,700 ± 1200 | 0.03 |
| (13-57)pS15pT18pS20 | 0.9 ± 0.5 | 22.2 | 81 ± 16 | 11.4 | 400 ± 140 | 24 | 3,700 ± 1,400 | 2.5 | 1,1800 ± 1,000 | 0.02 |
| (13-61)pS33pS37 | 2.0 ± 0.8 | 10.0 | 78 ± 23 | 11.8 | nm | — | nm | — | nm | — |
| (13-61)pS33pS46 | 1.4 ± 0.8 | 14.3 | 110 ± 20 | 8.4 | 2,400 ± 300 | 3.9 | 5,900 ± 800 | 1.6 | nm | — |
| (13-61)S33ApS37pS46 | 1.0 ± 0.6 | 16.0§ | 40 ± 15 | 25.0§ | 920 ± 100 | 3.3§ | nm | — | nm | — |
| (13-61)pS33pS37pS46 | 0.7 ± 0.6 | 28.6 | 32 ± 14 | 28.8 | 950 ± 280 | 9.9 | 4,600 ± 700 | 2.0 | nm | — |
| (13-61)S33A | 16 ± 2.1 | — | 1,000 ± 110 | — | 3,000 ± 100 | — | nm | — | nm | — |
nm, not measured.
*F = ratio Kd,unphosphorylated/Kd,phosphorylated
†Apparent values of Kd and F. Binding of the p53 TAD to KIX occurs in a competitive two-site binding mode (see text), and the reported dissociation constants are apparent values only.
‡The quoted uncertainty is the fitting error.
§Decrease in Kd relative to (13-61)S33A peptide.
In marked contrast to the CBP interactions, the effects of phosphorylation on binding of the p53 TAD to the N-terminal domain of HDM2 are highly specific (Table 1). In accord with previous results (11, 27), phosphorylation of Thr18 impairs binding by more than 20-fold. Triple-site phosphorylation at S15, T18, and S20 appears to inhibit binding even more.
The Effects of Multisite p53 Phosphorylation on CBP Binding Are Additive.
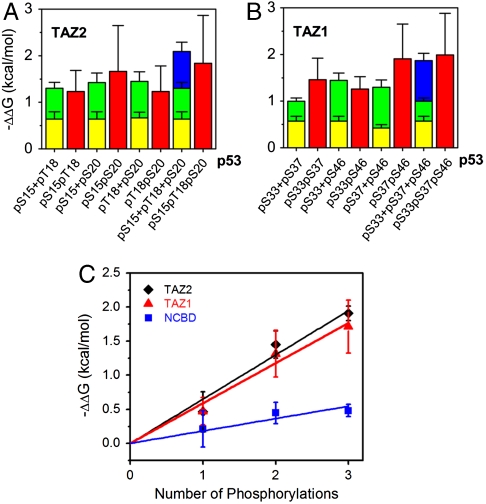
Successive phosphorylation events contribute in an additive manner to the free energy for binding of the p53 TAD to the CBP domains. Within experimental uncertainties, the free energies for binding of the doubly and triply phosphorylated peptides to the TAZ1 and TAZ2 domains are simply the sum of the binding free energies of the corresponding single-site phosphorylated peptides (Fig. 3 A and B). The average change in the free energy of binding (ΔΔG) to the TAZ2, TAZ1, and NCBD domains of CBP is plotted as a function of the number of phosphoryl groups in Fig. 3C. From the slope, each successive phosphoryl group on the p53 TAD enhances binding to the TAZ1 and TAZ2 domains by 0.6 kcal/mol, and by 0.2 kcal/mol for binding to the NCBD. Binding to KIX is also enhanced by ∼0.6 kcal/mol/phosphoryl group, but only for phosphorylation at S15, T18, and S20. Thus, an increase in the number of phosphoryl groups in the p53 TAD results in a proportional increase in binding affinity: p53 exhibits a graded response to phosphorylation, with the affinity for CBP domains dependent directly on the number of phosphoryl groups in the p53 TAD.
Fig. 3.
Additive effects of multisite p53 phosphorylation. (A) Differences in free energy [ΔΔG = -RT ln(Kd,unphosphorylated/Kd,phosphorylated)] for binding of single, double, and triple phosphorylated p53 peptides to TAZ2. Yellow, green, and blue bars indicate the effects of phosphorylation at single sites (pS15, pT18, or pS20). Red bars indicate the free energy differences associated with double or triple phosphorylation. (B) Differences in free energy (ΔΔG) for binding of single, double, and triple phosphorylated p53 peptides to TAZ1. Yellow, green, and blue bars indicate the effects of phosphorylation at single sites (pS33, pS37, or pS46). Red bars indicate the free energy differences associated with double or triple phosphorylation. (C) The average ΔΔG for single, double, or triple phosphorylation plotted as a function of the number of phosphoryl groups. The error bars represent the standard deviation of the average. The linear relationship implies additive effects of phosphorylation of p53 TAD for binding to the CBP domains.
Switch and Rheostat Models of Regulation.
Phosphorylation of p53 at Thr18 is a major negative effector of HDM2 binding, functioning as an on/off switch that controls the interaction between the p53 TAD and the N-terminal domain of HDM2 (9, 20). Mechanistically, Thr18 phosphorylation disrupts specific side chain interactions that stabilize the helical structure of the p53 TAD in the HDM2-bound state (28). In contrast, phosphorylation acts as a rheostat to regulate binding of the p53 TAD to CBP, with the affinity for the various CBP domains increasing in a graded manner as successive phosphoryl groups are added. Observation of a graded response provides important insights into the molecular basis for p53 activation and the mechanism by which p53 competes with cellular transcription factors for binding to CBP/p300.
Multisite phosphorylation adds another level of regulatory control and complexity to biological signaling (29, 30). Phosphorylation switches are commonly activated by phosphorylation of specific serine or threonine residues that induce conformational changes or modulate specific intermolecular hydrogen bonding interactions. Multisite phosphorylation events can function synergistically as on/off switches, or can function additively to regulate the biological response in a linear manner.
In the case of the p53 TAD, binding to HDM2 is controlled by a Thr18 phosphorylation switch; phosphorylation at other sites or multisite phosphorylation has no effect on HDM2 binding (Table 1). For interaction with the TAZ domains of CBP, however, the precise location of the phosphorylation site is not particularly important, but rather multiple phosphorylation events act additively to enhance binding to CBP domains in a linear manner (Fig. 3C). Hence, the effect of p53 phosphorylation on p53 binding to CBP can be attributed to bulk electrostatics, rather than site-specific intermolecular interactions. The TAZ domains of CBP are highly basic and have a large positive charge at neutral pH (+10.8 for TAZ1, +15.3 for TAZ2). These positively charged surfaces form favorable binding sites for amphipathic transcriptional activation domains that are rich in acidic residues and fold into helical structure upon binding (31, 32). The AD1 and AD2 motifs of the p53 TAD both contain acidic residues and form helices when bound to target proteins (33–35). Each phosphorylation event contributes two additional negative charges to the p53 TAD at pH 7, resulting in a higher net charge difference between the TAZ1/2 domains and the p53 TAD. The increased charge difference provides a stronger electrostatic driving force for intermolecular collisions and accelerates the association kinetics (36); the intrinsic disorder of the p53 TAD acts together with the favorable electrostatics to make binding highly efficient (37). The NCBD domain of CBP is more weakly electropositive (charge +5.7 at pH 7) and phosphorylation of the p53 TAD has a correspondingly smaller effect on binding affinity. The effects of phosphorylation on binding of the p53 TAD to the KIX domain are more complex due to the existence of multiple binding modes (26) and an apparent dependence on the location of the phosphoryl groups (Table 1). Because the TAZ1, TAZ2, and KIX domains of CBP and p300 have > 90% sequence identity and their overall charges are closely conserved, we expect that p53 will exhibit a similar rheostat response in binding to both CBP and p300.
There are nine phosphorylation sites in the human p53 TAD; however, only Ser15 is conserved across several species and Ser15, Thr18, and Ser20 across mammals (Fig. 1A). Phosphorylation sites are frequently found in disordered regions of proteins that are poorly conserved (38). The detailed patterns of multisite phosphorylation in the p53 TAD have not been fully delineated, although it has been observed that simultaneous phosphorylation of Ser15 and Ser20, or Thr18 and Ser20, has synergistic effects on the p53 response (22–24). Multisite phosphorylation creates dynamic regulatory networks that respond precisely and quantitatively to cellular signals and have the potential for complex information processing (30, 39). A network centered on variable multisite phosphorylation of p53 would be well adapted for sensing the nature and severity of cellular stress and for determining the cellular outcome—apoptosis or cell cycle arrest. Fine control by multisite phosphorylation has been observed previously; variable multisite phosphorylation serves as a rheostat to finely regulate DNA binding by Ets-1 and gating of the Kv2.1 potassium channel (40, 41). A primary function of multisite phosphorylation in intrinsically disordered proteins is to tune bulk electrostatic properties, leading to rheostat behavior or to ultrasensitive target binding once a threshold level of phosphorylation is reached (42, 43). It is worth noting that the average increase in binding energy contributed by each added phosphoryl group for binding of the p53 TAD to the CBP TAZ domains (∼0.6 kcal/mol) per phosphoryl is comparable to that for binding of Ets-1 to DNA (∼0.4 kcal/mol) per phosphoryl (40).
p53 Graded Response.
In vivo studies in NIH 3T3, C7, and B8 cells show that p53 protein levels are controlled in response to genotoxic stress and that increasing doses of stress agents result in a corresponding increase in p53 steady-state levels (44). There are two major events involved in p53 degradation and stability: negative regulation by HDM2, and positive regulation by CBP/p300 via acetylation of critical lysines (3). The interactions of p53 with HDM2 and CBP/p300 are both regulated by phosphorylation of the p53 TAD. However, our data suggest that, whereas phosphorylation of Thr18 acts as a simple binary switch to control binding to the N-terminal domain of HDM2, multisite phosphorylation of p53 plays an important role in regulating binding to CBP/p300 domains, functioning as a rheostat to enhance binding in a graded manner. Multisite phosphorylation provides a potential mechanistic explanation for the graded p53-dependent transcriptional response observed in certain cell types following genotoxic stress (44). The pattern and extent of phosphorylation is dependent upon cell type and the nature of the genotoxic stress (1, 2, 45, 46). Increases in phosphorylation levels have been correlated with increased or prolonged genotoxic stress, and hyperphosphorylation and hyperacetylation have been observed in some cancer cell lines (1, 2). The data reported in this work provide previously undescribed insights into the effect of multisite phosphorylation on the stabilization of p53 and recruitment of the CBP and p300 transcriptional coactivators.
Role of Multisite Phosphorylation in Regulation of the p53 Response.
CBP and p300 are central nodes in transcriptional regulatory networks in eukaryotes, and transcription factors must compete for binding to the limiting concentrations of CBP/p300 present in the cell (47, 48). For this reason, CBP/p300 activities must be fine-tuned for efficient and sufficient regulation of transcriptional processes.
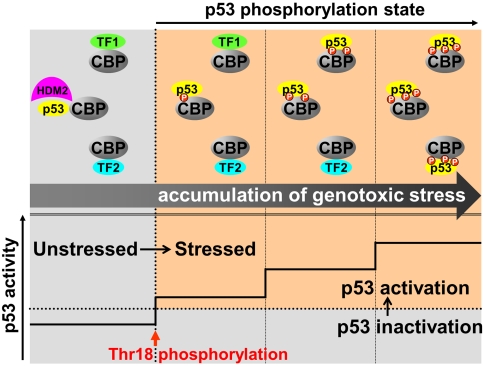
A model for the stabilization and activation of p53 by multisite phosphorylation is shown in Fig. 4. Single-site phosphorylation at Thr18 inhibits the interaction with the N-terminal region of HDM2, thereby stabilizing p53 against polyubiquitination (9) and relieving HDM2-mediated repression of the p21 gene (28). Mechanistically, activation of p21 expression is likely to occur by recruitment of a limited amount of CBP/p300, primarily through enhanced binding of the HDM2-free, Thr18-phosphorylated p53 TAD to TAZ2; the Kd decreases from 55 nM for binding of the p53:HDM2 complex to TAZ2 (28) to 6.5 nM for binding of the pThr18 TAD (Table 1). In contrast to interactions with TAZ2, addition of a single phosphoryl group to the p53 TAD enhances the interaction with other CBP/p300 domains (TAZ1, KIX, and NCBD) only slightly; binding remains too weak (Kd ranging from ∼400 nM to 10 μM) for p53 to compete effectively with cellular transcription factors for CBP/p300. Intense or prolonged genotoxic stress, leading to accumulation of p53 and phosphorylation of additional sites in the p53 TAD, will progressively enhance binding to the TAZ1, TAZ2, and KIX domains, thus ensuring increasingly efficient recruitment of the limited amounts of CBP/p300 and a graded p53 response. Mechanistically, the Nutlins function similarly to single-site Thr18 phosphorylation, competing off HDM2 even in the absence of phosphorylation (49) and activating transcription of p53-dependent genes through interactions with TAZ2 (Kd 20 nM).
Fig. 4.
Model for stabilization and activation of p53 by multisite phosphorylation. In unstressed cells, p53 forms a ternary complex with HDM2 and CBP/p300, thus promoting polyubiquitination and degradation of p53 (9). p53 must compete with cellular transcription factors (designated TF1 and TF2) for binding to limited amounts of CBP/p300 in cells. Upon induction of genotoxic stress, Thr18 of the p53 TAD is phosphorylated, lowering the affinity of p53 TAD for HDM2 and slightly increasing its affinity for the TAZ2, TAZ1, and KIX domains of CBP/p300. Phosphorylation of Thr18 functions as a switch in activation of p53. Prolonged or severe genotoxic stress leads to phosphorylation of additional sites in the p53 TAD, gradually increasing the affinity for the TAZ2, TAZ1, and KIX domains of CBP/p300 and allowing p53 to compete more effectively with cellular transcription factors for recruitment of CBP/p300.
The observation that multisite phosphorylation progressively enhances binding of the p53 TAD to the TAZ1 domain of CBP/p300 provides a plausible mechanistic explanation of the p53 response in hypoxia. Under mild to moderate hypoxia, the hypoxia inducible factor (HIF-1) accumulates and activates genes that are crucial for cell survival (50). Prolonged conditions of hypoxia or anoxia lead to stabilization of p53 and phosphorylation of the p53 TAD, which competes with the α-subunit of HIF-1 (HIF-1α) for binding to the TAZ1 domain of CBP/p300, resulting in repression of HIF-1 mediated transcription and activation of p53-regulated apoptotic genes (51–53). Because the C-terminal transactivation domain of HIF-1α binds to TAZ1 with a Kd of 10 nM (54), p53 cannot compete effectively until high levels of protein accumulate and until TAZ1 binding is enhanced by phosphorylation at multiple sites on the TAD. Inspection of Table 1 suggests that p53 must be phosphorylated at a minimum of two or three sites to lower the Kd into the 30- to 100-nM range, a level of phosphorylation that is likely reached only after prolonged or severe hypoxic stress.
The transactivation domains of p65 (RelA) and p53 also compete for binding to the TAZ1 domain of CBP/p300, leading to transcriptional cross-talk between NF-κB dependent cellular proliferation and survival pathways and p53-regulated apoptotic pathways (55, 56). By functioning as a molecular rheostat, multisite phosphorylation of the p53 TAD thus constitutes a possible mechanism for an exquisite level of control over critical cellular decisions, potentially contributing to life or death decisions over the fate of the cell.
Materials and Methods
Protein Expression and Purification.
The CBP TAZ1, TAZ2, KIX, and NCBD domains, the N-terminal domain of HDM2, and the p53(13-61) TAD were expressed and purified as described previously (9). p53(13-61)D57C and p53(13-61)S33A were prepared using site-directed mutagenesis and expressed and purified as for the wild-type protein. His6-tagged p38α and PRAK kinases were expressed in Escherichia coli BL21 (DE3) [DNAY] cells and purified by affinity chromatography on NiNTA resin (31). Details are given in SI Text.
Phosphorylation of p53.
Phosphorylated peptides were synthesized as described previously (9) or prepared biosynthetically. Phosphorylation at Ser33 and Ser46 was accomplished using p38α MAPK and Ser37 was phosphorylated using PRAK kinase. Double phosphorylation at Ser37 and Ser46 required a mutant p53 TAD (S33A) to prevent phosphorylation at Ser33. Details of the phosphorylation reactions are given in SI Text.
Determination of Kd by Fluorescence Anisotropy.
p53(13-61) D57C was labeled with Alexa Fluor 594 (Molecular Probes) using 5-fold molar excess of dye in 6 M guanidine hydrochloride, 50 mM Tris, pH 7.2 at room temperature for ∼3 h. The labeled protein was HPLC-purified by reverse phase chromatography and the mass verified by mass spectrometry. The affinities for binding of the phosphorylated p53 peptides to TAZ1 or TAZ2 were determined by a competition fluorescence anisotropy method. Changes in fluorescence anisotropy were monitored as phosphorylated p53 peptides were titrated into the complexes of dye-labeled p53(13-61)D57C with TAZ1 or TAZ2. A detailed description of the method and data analysis is given in SI Text.
Determination of Kd by NMR Titrations.
Measurement of Kd for the KIX and NCBD domains by fluorescence anisotropy was impractical because of the large quantity of phosphorylated p53 peptides needed to compete out the labeled p53 TAD due to the low binding affinities. Binding was therefore monitored using 1H-15N correlated NMR spectra, and Kd was determined from changes in the weighted average chemical shift differences Δδ(N,H)av = [(ΔδHN)2 + (ΔδN/5)2]1/2 assuming a one-site binding model (26). The one-site binding model assumes
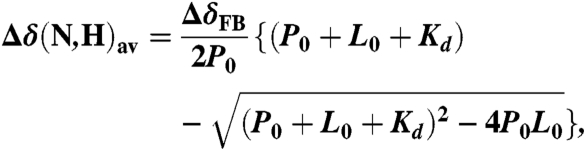 |
where ΔδFB is the chemical shift difference between the free and bound forms, and P0 and L0 are the total concentrations of KIX or NCBD and p53 peptides, respectively. The titration curves were fitted globally with an in-house fitting program nmrKd using the Levenberg–Marquardt algorithm (57).
Determination of Kd by ITC.
The affinities of the different phosphorylated p53 peptides for HDM2 were determined by ITC as described previously (9).
Supplementary Material
Acknowledgments.
We thank Euvel Manlapaz for technical support, Jane Dyson and Maria Martinez-Yamout for valuable discussions, Ashok Deniz for access to a fluorometer, and Peiqing Sun for supplying p38α and PRAK kinase plasmids. This work was supported by Grant CA96865 from the National Institutes of Health and by the Skaggs Institute for Chemical Biology. C.W.L was supported by Korea Research Foundation Grant KRF-2004-214-C00207 funded by the Korean Ministry of Education and Human Resource Development Basic Research Promotion Fund, and by a Skaggs training grant. J.C.F was supported by a Leukemia and Lymphoma Society Special Fellowship. A.C.M.F. was supported by a postdoctoral fellowship from the National Institute of Neurological Disorders and Stroke. M.A. was supported by a Grant-in-Aid for Scientific Research on Innovation Areas from MEXT, Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013078107/-/DCSupplemental.
References
- 1.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 2.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 6.Ayed A, et al. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 7.Dawson R, et al. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci USA. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman SR, et al. Polyubiquitination of p53 by a Ubiquitin Ligase Activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 11.Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene. 2009;28:2112–2118. doi: 10.1038/onc.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 13.Meek DW, Milne DM. Analysis of multisite phosphorylation of the p53 tumor-suppressor protein by tryptic phosphopeptide mapping. Methods Mol Biol. 2000;99:447–463. doi: 10.1385/1-59259-054-3:447. [DOI] [PubMed] [Google Scholar]
- 14.Dornan D, Hupp TR. Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300. EMBO Rep. 2001;2:139–144. doi: 10.1093/embo-reports/kve025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polley S, et al. Differential recognition of phosphorylated transactivation domains of p53 by different p300 domains. J Mol Biol. 2008;376:8–12. doi: 10.1016/j.jmb.2007.11.082. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins LMM, et al. Two distinct motifs within the p53 transactivation domain bind to the Taz2 domain of p300 and are differentially affected by phosphorylation. Biochemistry. 2009;48:1244–1255. doi: 10.1021/bi801716h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 18.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakaguchi K, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 20.Schon O, Friedler A, Freund S, Fersht AR. Binding of p53-derived ligands to MDM2 induces a variety of long range conformational changes. J Mol Biol. 2004;336:197–202. doi: 10.1016/j.jmb.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 21.Johnson TM, Attardi LD. Dissecting p53 tumor suppressor function in vivo through the analysis of genetically modified mice. Cell Death Differ. 2006;13:902–908. doi: 10.1038/sj.cdd.4401902. [DOI] [PubMed] [Google Scholar]
- 22.Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–2622. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jabbur JR, Zhang W. p53 Antiproliferative function is enhanced by aspartate substitution at threonine 18 and serine 20. Cancer Biol Ther. 2002;1:277–283. doi: 10.4161/cbt.81. [DOI] [PubMed] [Google Scholar]
- 24.Nakamizo A, et al. Phosphorylation of Thr18 and Ser20 of p53 in Ad-p53-induced apoptosis. Neuro-Oncology. 2008;10:275–291. doi: 10.1215/15228517-2008-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatti A, Li HH, Traugh JA, Liu X. Phosphorylation of human p53 on Thr-55. Biochemistry. 2000;39:9837–9842. doi: 10.1021/bi992454i. [DOI] [PubMed] [Google Scholar]
- 26.Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Mapping the interactions of the p53 transactivation domain with the KIX domain of CBP. Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 28.Jabbur JR, et al. Mdm-2 binding and TAF(II)31 recruitment is regulated by hydrogen bond disruption between the p53 residues Thr18 and Asp21. Oncogene. 2002;21:7100–7113. doi: 10.1038/sj.onc.1205856. [DOI] [PubMed] [Google Scholar]
- 29.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 30.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 31.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc Natl Acad Sci USA. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 34.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Lello P, et al. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein–protein association kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy Y, Onuchic JN, Wolynes PG. Fly-casting in protein-DNA binding: Frustration between protein folding and electrostatics facilitates target recognition. J Am Chem Soc. 2007;129:738–739. doi: 10.1021/ja065531n. [DOI] [PubMed] [Google Scholar]
- 38.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pufall MA, et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 41.Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2. 1 potassium channel by variable phosphorylation. Science. 2006;313:976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 42.Serber Z, Ferrell J. Tuning bulk electrostatics to regulate protein function. Cell. 2007;128:441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci USA. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joers A, Jaks V, Kase J, Maimets T. p53-dependent transcription can exhibit both on/off and graded response after genotoxic stress. Oncogene. 2004;23:6175–6185. doi: 10.1038/sj.onc.1207864. [DOI] [PubMed] [Google Scholar]
- 45.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 46.Boehme KA, Blattner C. Regulation of p53—insights into a complex process. Crit Rev Biochem Mol Biol. 2009;44:367–392. doi: 10.3109/10409230903401507. [DOI] [PubMed] [Google Scholar]
- 47.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 48.Kasper LH, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson T, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 50.Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- 51.Blagosklonny MV, et al. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 52.Schmid T, Zhou J, Köhl R, Brüne B. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1) Biochem J. 2004;380:289–295. doi: 10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammond EM, Giaccia AJ. The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun. 2005;331:718–725. doi: 10.1016/j.bbrc.2005.03.154. [DOI] [PubMed] [Google Scholar]
- 54.De Guzman RN, Martinez-Yamout M, Dyson HJ, Wright PE. Interaction of the TAZ1 domain of CREB-binding protein with the activation domain of CITED2: Regulation by competition between intrinsically unstructured ligands for non-identical binding sites. J Biol Chem. 2004;279:3042–3049. doi: 10.1074/jbc.M310348200. [DOI] [PubMed] [Google Scholar]
- 55.Wadgaonkar R, et al. CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J Biol Chem. 1999;274:1879–1882. doi: 10.1074/jbc.274.4.1879. [DOI] [PubMed] [Google Scholar]
- 56.Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in Fortran 77. The Art of Scientific Computing. Cambridge, UK: Cambridge Univ Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.