Abstract
As major consumers of heterotrophic bacteria and phytoplankton, microzooplankton are a critical link in aquatic foodwebs. Here, we show that a major marine microflagellate grazer is infected by a giant virus, Cafeteria roenbergensis virus (CroV), which has the largest genome of any described marine virus (≈730 kb of double-stranded DNA). The central 618-kb coding part of this AT-rich genome contains 544 predicted protein-coding genes; putative early and late promoter motifs have been detected and assigned to 191 and 72 of them, respectively, and at least 274 genes were expressed during infection. The diverse coding potential of CroV includes predicted translation factors, DNA repair enzymes such as DNA mismatch repair protein MutS and two photolyases, multiple ubiquitin pathway components, four intein elements, and 22 tRNAs. Many genes including isoleucyl-tRNA synthetase, eIF-2γ, and an Elp3-like histone acetyltransferase are usually not found in viruses. We also discovered a 38-kb genomic region of putative bacterial origin, which encodes several predicted carbohydrate metabolizing enzymes, including an entire pathway for the biosynthesis of 3-deoxy-d-manno-octulosonate, a key component of the outer membrane in Gram-negative bacteria. Phylogenetic analysis indicates that CroV is a nucleocytoplasmic large DNA virus, with Acanthamoeba polyphaga mimivirus as its closest relative, although less than one-third of the genes of CroV have homologs in Mimivirus. CroV is a highly complex marine virus and the only virus studied in genetic detail that infects one of the major groups of predators in the oceans.
Keywords: nucleocytoplasmic large DNA virus, horizontal gene transfer, viral evolution, DNA repair, 3-deoxy-D-manno-octulosonate
Predation by protistan grazers is a major pathway of carbon transfer and nutrient recycling in marine and freshwater systems (1); yet, viruses infecting phagotrophic protists in marine systems are largely unknown and completely unexplored genetically. The discovery of the giant Acanthamoeba polyphaga mimivirus in a freshwater amoeba, with its 1.2 million-base pair (bp) genome and 981 genes (2, 3), has sparked an intense debate about the biology and evolutionary origin of giant viruses. Whereas some researchers argue that giant viruses are “gene robbers” that have acquired their extensive gene collection by horizontal gene transfer (HGT) from cellular organisms (4–6), others favor the theory that these viruses date back to the emergence of eukaryotes and that most of their genes are viral in origin (7, 8). Recently, it has become evident that protists host the largest and most complex viruses known (9), that other giant viruses are likely widespread in oceans (10), and that some of these are pathogens of phytoplankton (11); yet, the only characterized giant viruses are those infecting species of Acanthamoeba. Ultimately, understanding the origin and evolution of giant viruses will be facilitated through the use of comparative genomics with other representative systems.
In this study, we used 454 pyrosequencing to sequence and de novo assemble the genome of a very large (300 nm capsid diameter) DNA virus, Cafeteria roenbergensis virus (CroV) strain BV-PW1, that was isolated from the coastal waters of Texas in the early 1990s (12). This lytic virus infects a marine heterotrophic flagellate, which is identical to C. roenbergensis strain VENT1 at the level of 18S rDNA. The host, which consumes bacteria and viruses (13), was originally misidentified as Bodo sp. (12). It is a 2-μm– to 6-μm–long bicosoecid heterokont phagotrophic flagellate (Stramenopiles) that is widespread in marine environments and is found in various habitats such as surface waters, deep sea sediments, and hydrothermal vents (14, 15). Populations of C. roenbergensis may be regulated by viruses in nature (16).
Results and Discussion
General Genome Features.
The genome of CroV is a linear double-stranded DNA molecule with a size of ≈730 kb, making this the second largest described viral genome. We sequenced and assembled the 618-kb central part of the viral chromosome, which is flanked on both ends by large and highly repetitive regions (Fig. 1). These terminal regions could potentially serve as protective caps for the protein-coding part of the genome, akin to telomeres in eukaryotes. The CroV genome is AT-rich (77% A+T), which is reflected in the distribution of codons and in the overall amino acid (aa) composition. AT-rich codons are consistently preferred over GC-rich ones, with the four most frequent aa (Lys, Ile, Asn, and Leu) each representing ≈10% of the overall aa (SI Appendix, Fig. S1).
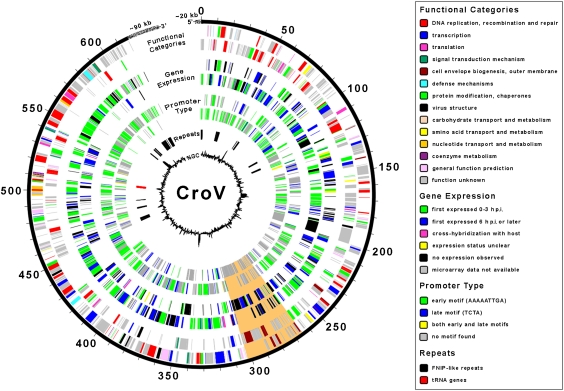
Fig. 1.
Genome diagram of CroV. Genome coordinates are given in kbs. Nested circles from outermost to innermost correspond to (i) predicted CDSs on forward strand and (ii) reverse strand; (iii) expression data for CDSs on forward strand and (iv) reverse strand; (v) gene promoter type for CDSs on forward strand and (vi) reverse strand; (vii) location of repetitive DNA elements; (viii) GC content plotted relative to the genomic mean of 23.35% G+C. The speckled regions at the chromosome ends are not drawn to scale and indicate terminal repeats for which no sequence information is available. A 38-kb genomic segment of putative bacterial origin is shaded orange.
Using conservative annotation criteria (SI Appendix), we identified 544 putative protein-coding sequences (CDSs) in the 618-kb central region of the CroV genome, which had a coding density of 90.1%. The average CDS was 1,025 nucleotides (nt) in length, and coding capacities ranged from 47 to 3,337 aa. Applying a BLASTP E-value cutoff of 1e-05, 267 CDSs (49%) displayed similarity to sequences in GenBank and 134 CDSs (25%) could be assigned to one or more Clusters of Orthologous Groups of proteins (COGs, E <0.001) (SI Appendix, Fig. S2). CroV CDSs and their annotations are listed in Dataset S1. Based on the distribution of top BLASTP hits, approximately one-half of the CroV genes displayed similarities to proteins found in eukaryotes, bacteria, archaea, and other giant viruses (SI Appendix, Fig. S3). Twenty-two percent of CroV CDSs had their top BLASTP hit among eukaryotes, but in the absence of genomic information about C. roenbergensis, no statement can be made about potential gene transfer between CroV and its host. Although most CroV CDSs were of unknown function, 32% of CDSs could be assigned a putative function and they provide insights into the biology of this giant virus. Several of these enzymatic functions have not been reported to be encoded by any other virus (SI Appendix, Table S1).
Translation Genes.
Viruses rely primarily on the protein translation apparatus of their hosts; it is therefore unusual to find viral genes associated with protein synthesis. CroV encodes an isoleucyl-tRNA synthetase and putative homologs of eukaryotic translation initiation factors eIF-1, eIF-2α, eIF-2β/eIF-5, eIF-2γ, eIF-4AIII, eIF-4E, and eIF-5B. Using the transfer RNA gene prediction software tRNAScan-SE, we identified 22 tRNA genes, clustered in a 2.8-kb region around position 510,000 (Fig. 1 and SI Appendix, Table S2). We also found two putative tRNA-modifying enzymes in CroV, tRNA pseudouridine 5S synthase and tRNAIle lysidine synthetase. These genes add to a rapidly growing number of virus-encoded protein translation components. Some tRNA genes are scattered among bacteriophages and eukaryotic viruses such as the phycodnaviruses (17, 18), and four tRNA synthetases along with several putative translation factors are found in Mimivirus (2). These findings imply that CroV and similarly complex viruses encode genes to modify and regulate the host translation system to their own advantage, which results in a “lifestyle” that is less dependent on host cell components than that of smaller viruses.
DNA Repair Genes.
The ability to repair various kinds of DNA damage is well documented among large DNA viruses (19, 20). Given that the AT-rich genome of CroV is exposed to high solar irradiance in surface waters of the ocean and is therefore likely to suffer from DNA lesions such as pyrimidine dimers, it is not surprising that CroV encodes multiple DNA repair proteins. We found putative components of several DNA repair mechanisms, including a presumably complete base excision repair pathway with formamidopyrimidine DNA glycosylase, a family 1 apurinic/apyrimidinic (AP) endonuclease, a family X DNA polymerase, and an NAD-dependent DNA ligase. Further DNA repair proteins include DNA mismatch repair protein MutS, XPG endonuclease, a homolog of the alkylated DNA repair protein AlkB, and two DNA photolyases. Photolyases are classified into three major groups: Cyclobutane pyrimidine dimer (CPD) photolyases, (6-4) photolyases, and single-stranded DNA photolyases (ref. 21 and references therein). The CPD photolyases are further subdivided into class I and class II enzymes, the former being more prevalent in bacteria and the latter more frequent in eukaryotes. The gene product of crov115 is a predicted CPD class I photolyase and represents the first viral homolog in this class (SI Appendix, Figs. S4 and S5). The second CroV photolyase (crov149) does not belong to any of the established types of photolyases. Instead, it is related to a recently described group of photolyases/cryptochromes that are present in several bacterial phyla and the euryarchaeotes (21) (SI Appendix, Figs. S4 and S6). The only eukaryotic member in this group (Paramecium tetraurelia) is also the closest homolog to the CroV and Mimivirus sequences and may have acquired this gene by HGT from a giant virus (SI Appendix, Fig. S6).
Transcription Genes.
Large DNA viruses typically carry hundreds of genes, including several that regulate gene expression. Among the predicted transcriptional genes in CroV are eight DNA-dependent RNA polymerase II subunits, at least six transcription factors involved in transcription initiation, elongation, and termination, a trifunctional mRNA capping enzyme, a poly(A) polymerase, and several helicases. The complex transcriptional machinery encoded by CroV suggests that viral gene transcription does not depend on host enzymes and likely occurs in the cytoplasm. Interestingly, CroV contains a CDS with high similarity to an ELP3-like histone acetyltransferase (HAT, COG1243, 2e-46), a gene previously not seen in viruses. In combination with other unidentified viral gene products, the CroV HAT may enable the virus to directly modulate the genome condensation state of the host and, thus, exert control over its transcriptional activity. Alternatively, this enzyme may be involved in replication and packaging of the virus genome itself. Another unusual characteristic of CroV is the presence of three DNA topo-isomerase (Topo) genes of types IA, IB, and IIA. TopoIA and TopoIIA are very similar to their counterparts in Mimivirus, and HGT events from bacteria (eventually via a eukaryotic phagotrophic host) have been proposed for these genes (22). CroV TopoIB is the first viral homolog of the eukaryotic subfamily, whereas the TopoIB encoded by Mimivirus falls within the bacterial group (SI Appendix, Fig. S7) and is functionally more similar to the poxvirus enzymes (23). Despite apparently different evolutionary trajectories, the presence of three Topo genes in CroV and Mimivirus suggests a crucial role for these enzymes in transcription, replication, or packaging of giant virus genomes.
Repetitive DNA and Ubiquitin Components.
Approximately 5% of the genome (excluding the terminal regions) consisted of repetitive elements. The most prevalent was a 22-aa–long leucine-rich repeat similar to the FNIP/IP22 repeat (Pfam entry PF05725) that had >400 copies in the CroV genome and was present in at least 28 CDSs (Fig. 1 and Dataset S1). This repeat also occurs in Mimivirus and Dictyostelium discoideum (24). Whereas leucine-rich repeats are known to mediate protein–protein interactions in a variety of proteins with diverse functions (25), the role of these repeats in CroV is unknown. In Mimivirus, FNIP/IP22 repeat-containing genes also possess an N-terminal F-box domain, which mediates interaction with the ubiquitin (Ub) pathway (26). Ub signaling appears to be a general strategy used by nucleocytoplasmic large DNA viruses (NCLDVs) to counter host defenses, because multiple Ub-conjugating and Ub-hydrolyzing enzymes have been found in these viruses (26). Furthermore, it has been shown that orthopoxvirus replication requires a functional Ub-proteasome system (27). In CroV, we identified a small arsenal of genes encoding proteins predicted to function in the Ub pathway, including an E1 Ub-activating enzyme, six E2 Ub-conjugating enzymes, two deubiquinating enzymes, and one Ub gene. The specific means of how CroV and other giant viruses use Ub signaling to interact with their hosts remain to be determined.
CroV Harbors Four Inteins.
No introns were detected in the genome, but four CDSs contained an intein, i.e., a self-splicing protein sequence inserted in highly conserved regions of a host protein (28). All four CroV inteins are part of NCLDV core genes that are thought to play a key role in DNA replication and transcription: DNA-dependent DNA polymerase B (PolB), TopoIIA, DNA-dependent RNA polymerase II subunit 2 (RPB2), and the large subunit of ribonucleotide reductase (RNR). Ten other inteins have been found in viruses infecting eukaryotes (29), including PolB inteins in Mimivirus (30), Heterosigma akashiwo virus (31), and Chrysochromulina ericina virus (32) as well as RNR inteins in four iridoviruses and the chlorella virus NY-2A (33). With the exception of a gene fragment from Emiliania huxleyi virus 163 (34), the CroV RPB2 intein constitutes the only viral report of an intein in RPB2. Finally, the CroV TopoIIA intein is a unique case of an intein in a DNA topoisomerase II gene, thus extending the known range of intein-containing genes. All four CroV inteins possess the conserved nucleophilic residues that are required for the standard splicing reaction [C/S at the N-terminal splice junction and N(C/S/T) at the C-terminal splice junction] (28) and are therefore probably capable of autocatalytic excision.
Microarray Analysis.
A microarray experiment was undertaken to determine which CroV genes were unambiguously transcribed in infected cells and if there was a clear temporal pattern in the transcription of those genes. We detected viral transcripts in infected C. roenbergensis cells by fluorescently labeling mRNA isolated at different time points during the infection cycle, which lasted 12–18 h in C. roenbergensis strain E4-10. We then hybridized the labeled transcripts to glass slides spotted with oligonucleotide probes for 438 of the 544 predicted CroV genes (SI Appendix). Detectable levels of expression were found for 274 genes (63%), 152 genes (35%) were below the detection limit, 4 (1%) cross-hybridized with host mRNA isolated from uninfected cells, and 8 (2%) could not be assigned a clear on/off status (Fig. 1 and Dataset S1). Therefore, approximately one-half of the predicted genes and 63% of the genes we tested were expressed during infection under our laboratory conditions. This percentage is comparable with the observed expression of 65% of viral genes during infection of the marine phytoplankter Emiliania huxleyi by EhV-86 (35). However, recent gene expression studies in PBCV-1 and Mimivirus validated transcription for nearly all of their predicted genes (3, 36). It seems therefore likely that our microarray data underestimated the true extent of transcriptional activity in CroV. All of the previously mentioned translation-related genes in CroV, as well as most of the “virus-atypical” genes were expressed (Dataset S1), suggesting that these genes are functional. Although the microarray experiment was designed primarily to validate CroV gene predictions and cannot be exploited quantitatively, the data allowed us to recognize some general trends of CroV gene expression. Based on the time points at which transcripts were first detected, we could distinguish between an early and a late phase of CroV gene expression. The early phase lasted from 0 h to 3 h after infection (h p.i.) and affected 150 genes. The majority of DNA replication and transcription genes belonged to the early class. The late phase was characterized by genes that were first detected in the microarray at 6 h p.i. or later. The 124 genes in this class included all of the predicted structural components, such as the major and minor capsid proteins. Further and more extensive analysis of the CroV transcriptome may be able to refine this preliminary temporal classification.
Promoter Analysis.
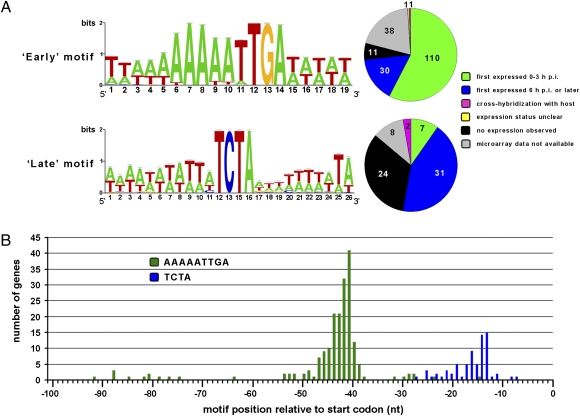
The intergenic regions had an average size of 71 ± 64 bp. We examined the 100-nt region upstream of the predicted start codons for possible promoter motifs by using MEME software (37). A perfectly conserved “AAAAATTGA” motif, flanked by AT-rich sequences, was found to precede 127 CroV CDSs (23%) (Fig. 2A). The MEME E-value for this motif was 9e-170. Allowing one mismatch per sequence at the less strongly conserved positions one to six of the AAAAATTGA motif increased the number of positive CDSs to 191 (35%). The majority of CDSs that displayed this motif in their immediate upstream region belonged to the “early” temporal category (Fig. 2A). We therefore classified this motif as an early gene promoter in CroV. Our results are in agreement with findings from Mimivirus, where a nearly identical early promoter motif (AAAATTGA) is associated with 45% of Mimivirus genes (3, 38). But, in contrast to Mimivirus, where the motif is found preferentially in the −50 to −110 region, the early promoter motif in CroV displayed a much narrower distribution, with a peak at position −40 relative to the predicted start codon (Fig. 2B).
Fig. 2.
Early and late gene promoter motifs in CroV. (A) Sequence logos depicting the consensus sequence for putative early (AAAAATTGA) and late (TCTA) promoter motifs. Pie charts show gene expression data for those CDSs that contained the respective motifs within their immediate 5′ upstream regions. The majority of CDSs associated with the AAAAATTGA motif were first seen expressed at 0–3 h p.i., whereas transcripts for most of the TCTA-associated CDSs were not detected until 6 h p.i. or later. (B) Positional distribution of the two motifs relative to the predicted start codon. A narrow distribution with a peak around position −40 is observed for the AAAAATTGA motif (n = 191). The TCTA motif (n = 72) occurs preferentially at position −13 to −21. The search for this motif was restricted to the upstream 30-nt region.
We then searched for a possible late promoter motif starting with a representative set of six CDSs, all predicted to encode capsid components (SI Appendix). Five of the six genes exhibited the conserved tetramer “TCTA,” flanked by AT-rich regions on either side, in their −11 to −20 region (Fig. 2). Based on this profile, we expanded the search to all CroV CDSs and identified 72 that were positive for the TCTA motif signature. A MEME search on the 30-nt upstream region of the 124 genes classified as “late” yielded a very similar motif (MEME E-value 5e-04; SI Appendix, Fig. S8). As shown in Fig. 2A, most CDSs with the TCTA promoter motif were first expressed at 6 h p.i. or later, supporting our conclusion that this sequence motif represents a promoter element for genes transcribed during the late phase of CroV infection. The CroV late promoter motif is unrelated to the putative late promoter motif identified in Mimivirus (3).
A Thirty-Eight–Kilobase Genomic Fragment Involved in Carbohydrate Metabolism.
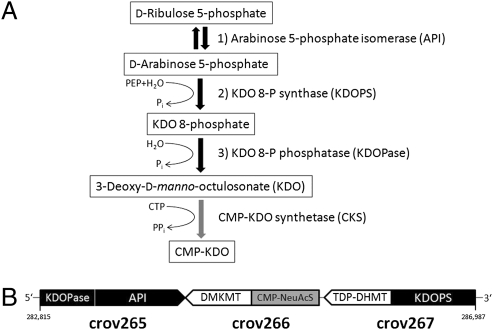
Upon examination of the CroV promoter distribution, we noticed that neither early not late promoter motifs were associated with CDSs located between the genomic positions 264,800 and 302,500 (Fig. 1). Of these 34 CDSs (crov242–crov275), 14 were most similar to bacterial proteins (SI Appendix, Table S3; BLASTP E-value <1e-05) and 7 of them are predicted to function in carbohydrate metabolism (Dataset S1). Among them, we identified enzymes for the biosynthesis of 3-deoxy-d-manno-octulosonate (KDO) (Fig. 3). In Gram-negative bacteria, KDO is an essential core component of the lipopolysaccharide layer, linking lipid A to polysaccharides (39). Biosynthesis of KDO, which is also found in the green alga Chlorella and the cell wall of higher plants, involves the three enzymes arabinose 5-phosphate isomerase (API), KDO 8-phosphate synthase (KDOPS), and KDO 8-phosphate phosphatase (KDOPase). A cytidylyltransferase (CMP-KDO synthetase, CKS) is then required to activate KDO for downstream reactions (Fig. 3A). We identified in crov265 a bifunctional KDOPase/API and in the N-terminal domain of crov267 a KDOPS homolog. The C-terminal domain of crov267 is a predicted dTDP-6-deoxy-l-hexose 3-O-methyltransferase, and crov266 encodes a predicted bifunctional N-acylneuraminate cytidylyltransferase (CMP-NeuAcS)/demethylmenaquinone methyltransferase (Fig. 3B and SI Appendix, Figs. S9–S12). Whether the cytidylyltransferase in crov266 is a functional CMP-NeuAcS and accordingly involved in sialic acid activation, as suggested by phylogenetic analysis (SI Appendix, Fig. S12), or rather a structurally related KDO-activating CKS, remains to be tested. The remaining CroV CDSs with functional annotation in this region are predicted glycosyltransferases and other sugar-modifying enzymes (Dataset S1). The presence of these genes and the finding that 10 of them were expressed (Dataset S1) suggests a role in viral glycoprotein biosynthesis and that the virion surface may be coated with KDO- or sialic acid-like glycoconjugates, which could be involved in virion-cell recognition. Given that the CDSs in this region lack the early/late promoter signals and have no homologs in Mimivirus, the 38-kb region must have been acquired after the CroV lineage split from the Mimivirus lineage. Because many of the CDSs in this region were most similar to bacterial genes (SI Appendix, Figs. S9 and S12 and Table S3), it is tempting to speculate that they may have been acquired from a bacterium, considering that CroV frequently encounters phagocytosed bacteria inside the host cytoplasm and encodes several enzymes that might catalyze an integration of foreign DNA (e.g., transposase crov356). Genomic islands of putative bacterial origin have been identified in other giant viruses such as phycodnaviruses and Mimivirus, but in contrast to CroV, these bacterial gene clusters tend to be located toward the ends of the linear viral chromosomes (40). However, given that the GC content of the 38-kb region is even lower than that of the rest of the CroV genome (19.4% vs. 23.6% G+C) and that some of these proteins occupy a phylogenetic position between bacterial and eukaryotic homologs (e.g., KDOPS and KDOPase; SI Appendix, Figs. S10 and S11), we cannot rule out alternative scenarios for the origin of this region.
Fig. 3.
The predicted KDO biosynthesis pathway in CroV. (A) Schematic of the three enzymatic steps that transform d-ribulose 5-phosphate into KDO. Activation of KDO to CMP-KDO is catalyzed by the cytidylyltransferase CKS. PEP, phosphoenolpyruvate. (B) Organization of the predicted KDO gene cluster (crov265–crov267) in the CroV genome. All three CDSs are predicted bifunctional enzymes. Genome coordinates are given. DMKMT, demethylmenaquinone methyltransferase; TDP-DHMT, dTDP-6-deoxy-l-hexose 3-O-methyltransferase; CMP-NeuAcS, N-acylneuraminate cytidylyltransferase.
Phylogenetic Relationship.
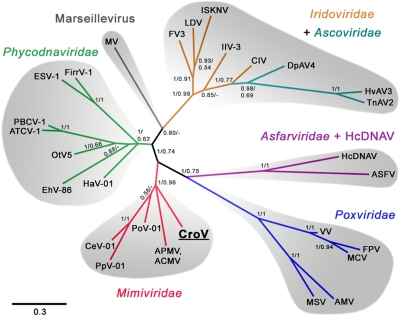
Based on the presence and phylogenetic analysis of a set of core genes (SI Appendix, Fig. S13), CroV is an addition to the presumably monophyletic group of NCLDVs (2, 26, 41), which includes the families Ascoviridae, Asfarviridae, Iridoviridae, Mimiviridae, Phycodnaviridae, Poxviridae, and the newly discovered Marseillevirus (42). In a recent study by Yutin et al. (43), genes encoded by NCLDVs were categorized into groups that presumably evolved from a common ancestor and subsequently diversified in the various NCLDV families. Using this dataset of Nucleo-Cytoplasmic Virus Orthologous Genes (NCVOGs), we found that at least 172 CroV CDSs belonged to an existing NCVOG (Dataset S1). Thirty-two percent of CroV CDSs were significantly similar to a Mimivirus gene (any Mimivirus hit with a BLASTP E-value >1e-05) and 22 CroV CDSs had their only detectable GenBank homolog in Mimivirus. CroV therefore appears to be the closest known relative to Mimivirus, despite large differences in genome (730 kb vs. 1,181 kb) and capsid size (300 nm vs. 500 nm). The CroV–Mimivirus relationship was further corroborated by phylogenetic analysis of PolB, a commonly used marker gene to infer phylogenetic relationships among NCLDVs. Bayesian Inference analysis of PolB resulted in a strongly supported clade comprising the largest known viruses: Mimivirus, CroV, and three partially sequenced viruses infecting the marine microalgae Phaeocystis pouchetii (PpV), Chrysochromulina ericina (CeV), and Pyramimonas orientalis (PoV) (Fig. 4). These three algal viruses, for which only PolB and major capsid protein (MCP) sequences are available, also possess very large DNA genomes (485 kb, 510 kb, and 560 kb, respectively) and are proposed members of the family Phycodnaviridae, although a taxonomic revision of this tentative assignment has been proposed (10). Similarly, when the MCP was used to reconstruct the NCLDV phylogeny (SI Appendix, Fig. S14), these five viruses formed a monophyletic group that also included Heterosigma akashiwo virus, another large DNA virus that is assigned to the Phycodnaviridae. The topology of the NCLDV tree strongly suggests that the five largest viral genomes are more closely related to each other than to other NCLDV families and that they may have originated from a relatively recent ancestral virus that must have already been a bona fide NCLDV with a very large genome, probably encoding >150 proteins.
Fig. 4.
Phylogenetic reconstruction of NCLDV members. The unrooted Bayesian Inference (BI) tree was generated from a 263-aa alignment of conserved regions of DNA polymerase B. Intein insertions were removed before alignment. Nodes are labeled with BI posterior probabilities and maximum likelihood bootstrap values (500 replicates). Abbreviations and accession numbers (GenBank unless stated otherwise) are as follows: ACMV, Acanthamoeba castellanii mamavirus, from ref. 43; AMV, Amsacta moorei entomopoxvirus, NP_064832; APMV, A. polyphaga mimivirus, YP_142676; ASFV, African swine fever virus, NP_042783; ATCV-1, Acanthocystis turfacea chlorella virus 1, YP_001427279; CeV-01, C. ericina virus 01, ABU23716; CIV, Chilo iridiscent virus, NP_149500; CroV, C. roenbergensis virus; DpAV4, Diadromus pulchellus ascovirus 4a, CAC19127; EhV-86, E. huxleyi virus 86, YP_293784; ESV-1, Ectocarpus siliculosus virus 1, NP_077578; FirrV-1, Feldmannia irregularis virus 1, AAR26842; FPV, Fowlpox virus, NP_039057; FV3, Frog virus 3, YP_031639; HaV-01, H. akashiwo virus 01, BAE06251; HcDNAV, Heterocapsa circularisquama DNA virus, DDBJ accession no. AB522601; HvAV3, Heliothis virescens ascovirus 3e, YP_001110854; IIV-3, Invertebrate iridiscent virus 3, YP_654692; ISKNV, Infectious spleen and kidney necrosis virus, NP_612241; LDV, Lymphocystis disease virus, YP_073706; MCV, Molluscum contagiosum virus, AAL40129; MSV, Melanoplus sanguinipes entomopoxvirus, NP_048107; MV, Marseillevirus, MAR_ORF329, GU071086; OtV5, Ostreococcus tauri virus 5, YP_001648316; PBCV-1, P. bursarium chlorella virus 1, NP_048532; PoV-01, P. orientalis virus 01, ABU23717; PpV-01, P. pouchetti virus 01, ABU23718; TnAV2, Trichoplusia ni ascovirus 2c, YP_803224; VV, Vaccinia virus, AAA98419.
Conclusions
We present here the genetic analysis of a virus infecting a marine phagotroph. With a genome size larger than that of some cellular organisms, CroV is an example of an extraordinarily complex virus. It possesses a large number of predicted genes involved in DNA replication, transcription, translation, protein modification, and carbohydrate metabolism, indicating that CroV has a highly autonomous propagation strategy during infection.
The mechanisms by which such enormous virus genomes evolved have been much discussed (40, 44, 45). Most studies have focused on Mimivirus, because it represents the most extreme case of a giant virus and is the largest dataset available. The majority of Mimivirus genes have no cellular homologs and are presumably very ancient (46), up to one-third of its genes arose through gene and genome duplication (45), and <15% of Mimivirus genes may have been horizontally transferred from eukaryotes and bacteria (6). Our analysis of the CroV genome is consistent with this general picture of giant virus genome evolution. Gene duplication and lineage-specific expansion of the FNIP/IP22 repeat are two factors that clearly contributed to the enormous size of the CroV genome. Examples of duplicated genes are the paralogous groups of CDSs crov027–crov031 (contain FNIP/IP22 repeats), crov420–crov422 (unknown function), and some of the tRNA genes. A potential case of large-scale HGT from a bacterium is represented by the 38-kb genomic segment that differs in coding content and promoter regions from the rest of the viral genome. The remaining CDSs with cellular homologs are more difficult to categorize, because genes can be transferred from cells to viruses and vice versa. However, the majority of CroV CDSs show no significant similarity to any sequences in the public databases and their evolutionary origin remains hidden.
The array of “organismal” genes found in CroV further closes the overlap in metabolic coding capacity between large viruses and cellular life forms. This continued blurring of the distinction between what is considered living and nonliving adds to the ongoing debate about the puzzling evolutionary history of giant viruses (7, 8, 44). Moreover, the PolB gene of CroV has high similarity with those of other marine virus isolates, relatives of which appear to be widespread in the oceans (10), suggesting that CroV represents a major group of largely unknown but ecologically important marine viruses.
Materials and Methods
Flagellate Growth and Virus Purification.
C. roenbergensis strain E4-10 was isolated from coastal waters near Yaquina Bay, OR, as described (13).
Cultures of C. roenbergensis were grown in f/2-enriched seawater medium supplemented with 0.01% (wt/vol) yeast extract to stimulate bacterial growth. The mixed assembly of bacteria in the cultures served as the food source for C. roenbergensis. Cultures were kept at room temperature (≈22 °C) in the dark. Typically, 1-L plastic Erlenmeyer flasks containing 250 mL of exponentially growing C. roenbergensis were infected at a cell density of 5×104 cells per mL by adding 100 μL (multiplicity of infection ≈0.5) of crude CroV-containing lysate. CroV purification is described in SI Appendix.
Genome Sequencing and Assembly.
Phenol-chloroform extracted genomic DNA was sequenced by 454 pyrosequencing on GS 20 and GS FLX platforms. The two datasets were assembled individually and resulting contigs were analyzed with Sequencher (Gene Codes). Gap closing was achieved by a combination of multiplex PCR, bioinformatic prediction methods followed by PCR verification and sequencing, and a genomic shotgun library using the pSMART vector (Lucigen).
SI Appendix contains further details and experimental procedures on genome annotation and microarray analysis.
Supplementary Material
Acknowledgments
We thank D. R. Garza, T. St. John, and A. S. Lang for help with the virus purification protocol during an early phase of the project, A. I. Culley and R. Adelshin for advice and discussion on DNA cloning and sequencing issues, A. M. Chan for assistance with flagellate culturing, and the staff at the Liverpool Microarray Facility for microarray fabrication. This work was supported by the Natural Science and Engineering Research Council of Canada Discovery Grants Program (to C.A.S.), the Tula Foundation through the Centre for Microbial Diversity and Evolution (C.A.S.), Natural Environment Research Council (NERC) Environmental Genomics Thematic Program Grants NE/A509332/1, NE/D001455/1WHW (to W.H.W.), and NERC SPG/MGF170 (to M.J.A.), and fellowships awarded by the Gottlieb Daimler- and Karl Benz-Foundation, Germany, and the University of British Columbia (to M.G.F.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. GU244497 (CroV genome) and GU249597 (partial 18S sequence from C. roenbergensis strain E4-10)]. The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE19051).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007615107/-/DCSupplemental.
References
- 1.Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Legendre M, et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–674. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filée J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreira D, López-García P. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 6.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12–21. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Etten JL, Lane LC, Dunigan DD. DNA viruses: The really big ones (giruses) Annu Rev Microbiol. 2010;64:83–99. doi: 10.1146/annurev.micro.112408.134338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monier A, et al. Marine mimivirus relatives are probably large algal viruses. Virol J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandaa RA, Heldal M, Castberg T, Thyrhaug R, Bratbak G. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae) Virology. 2001;290:272–280. doi: 10.1006/viro.2001.1161. [DOI] [PubMed] [Google Scholar]
- 12.Garza DR, Suttle CA. Large double-stranded DNA viruses which cause the lysis of a marine heterotrophic nanoflagellate (Bodo sp) occur in natural marine viral communities. Aquat Microb Ecol. 1995;9:203–210. [Google Scholar]
- 13.Gonzalez JM, Suttle CA. Grazing by marine nanoflagellates on viruses and virus-sized particles: Ingestion and digestion. Mar Ecol Prog Ser. 1993;94:1–10. [Google Scholar]
- 14.Scheckenbach F, Wylezich C, Weitere M, Hausmann K, Arndt H. Molecular identity of strains of heterotrophic flagellates isolated from surface waters and deep-sea sediments of the South Atlantic based on SSU rDNA. Aquat Microb Ecol. 2005;38:239–247. [Google Scholar]
- 15.Atkins MS, Teske AP, Anderson OR. A survey of flagellate diversity at four deep-sea hydrothermal vents in the Eastern Pacific Ocean using structural and molecular approaches. J Eukaryot Microbiol. 2000;47:400–411. doi: 10.1111/j.1550-7408.2000.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 16.Massana R, del Campo J, Dinter C, Sommaruga R. Crash of a population of the marine heterotrophic flagellate Cafeteria roenbergensis by viral infection. Environ Microbiol. 2007;9:2660–2669. doi: 10.1111/j.1462-2920.2007.01378.x. [DOI] [PubMed] [Google Scholar]
- 17.Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007;17:1486–1495. doi: 10.1101/gr.6649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv Virus Res. 2006;66:293–336. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan V, Schnitzlein WM, Tripathy DN. Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus. J Virol. 2001;75:1681–1688. doi: 10.1128/JVI.75.4.1681-1688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuta M, et al. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl Environ Microbiol. 1997;63:1551–1556. doi: 10.1128/aem.63.4.1551-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas-Lledó JI, Lynch M. Evolution of mutation rates: Phylogenomic analysis of the photolyase/cryptochrome family. Mol Biol Evol. 2009;26:1143–1153. doi: 10.1093/molbev/msp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Benarroch D, Claverie JM, Raoult D, Shuman S. Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria. J Virol. 2006;80:314–321. doi: 10.1128/JVI.80.1.314-321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Day DH, Suhre K, Myre MA, Chatterjee-Chakraborty M, Chavez SE. Isolation, characterization, and bioinformatic analysis of calmodulin-binding protein cmbB reveals a novel tandem IP22 repeat common to many Dictyostelium and Mimivirus proteins. Biochem Biophys Res Commun. 2006;346:879–888. doi: 10.1016/j.bbrc.2006.05.204. [DOI] [PubMed] [Google Scholar]
- 25.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 26.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Teale A, et al. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J Virol. 2009;83:2099–2108. doi: 10.1128/JVI.01753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Inteins: Structure, function, and evolution. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 29.Perler FB. InBase: The Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata H, Raoult D, Claverie JM. A new example of viral intein in Mimivirus. Virol J. 2005;2:8. doi: 10.1186/1743-422X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagasaki K, Shirai Y, Tomaru Y, Nishida K, Pietrokovski S. Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl Environ Microbiol. 2005;71:3599–3607. doi: 10.1128/AEM.71.7.3599-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen JB, Larsen A, Bratbak G, Sandaa RA. Phylogenetic analysis of members of the Phycodnaviridae virus family, using amplified fragments of the major capsid protein gene. Appl Environ Microbiol. 2008;74:3048–3057. doi: 10.1128/AEM.02548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald LA, et al. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007;358:472–484. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin TJ, Butler MI, Poulter RT. Multiple, non-allelic, intein-coding sequences in eukaryotic RNA polymerase genes. BMC Biol. 2006;4:38–54. doi: 10.1186/1741-7007-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson WH, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 36.Yanai-Balser GM, et al. Microarray analysis of Paramecium bursaria chlorella virus 1 transcription. J Virol. 2010;84:532–542. doi: 10.1128/JVI.01698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 38.Suhre K, Audic S, Claverie JM. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci USA. 2005;102:14689–14693. doi: 10.1073/pnas.0506465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raetz CR. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 40.Filée J, Chandler M. Convergent mechanisms of genome evolution of large and giant DNA viruses. Res Microbiol. 2008;159:325–331. doi: 10.1016/j.resmic.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer M, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filée J, Siguier P, Chandler M. I am what I eat and I eat what I am: Acquisition of bacterial genes by giant viruses. Trends Genet. 2007;23:10–15. doi: 10.1016/j.tig.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata H, Claverie JM. Unique genes in giant viruses: Regular substitution pattern and anomalously short size. Genome Res. 2007;17:1353–1361. doi: 10.1101/gr.6358607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.