Abstract
The N-glycosidic bond can be hydrolyzed spontaneously or by glycosylases during removal of damaged bases by the base excision repair pathway, leading to the formation of highly mutagenic apurinic/apyrimidinic (AP) sites. Organisms encode for evolutionarily conserved repair machinery, including specific AP endonucleases that cleave the DNA backbone 5′ to the AP site to prime further DNA repair synthesis. We report on the DNA polymerase X from the bacterium Bacillus subtilis (PolXBs) that, along with polymerization and 3′–5′-exonuclease activities, possesses an intrinsic AP-endonuclease activity. Both, AP-endonuclease and 3′–5′-exonuclease activities are genetically linked and governed by the same metal ligands located at the C-terminal polymerase and histidinol phosphatase domain of the polymerase. The different catalytic functions of PolXBs enable it to perform recognition and incision at an AP site and further restoration (repair) of the original nucleotide in a standalone AP-endonuclease-independent way.
Keywords: apurinic/apyrimidinic-lyase, site-directed mutagenesis
Genomes are continuously insulted by exogenous and endogenous genotoxic agents, as ionizing radiation, drugs, and (by)products of normal cellular metabolism that generate reactive oxygen species (ROS) leading to mainly nonbulky DNA lesions (1). Base excision repair (BER) is the major pathway involved in the removal of this type of damage, and its importance for cell survival is reflected by its conservation from bacteria to eukaryotes (2). During the first steps of BER, highly mutagenic apurinic/apyrimidinic (AP) intermediates are produced as a result of hydrolytic cleavage of the altered base-sugar bond by mono- (class II) and/or bifunctional (class I) DNA N-glycosylases (ref. 3 and references therein), or from spontaneous DNA base loss, causing replication and transcription inhibition if left unrepaired (4, 5). AP endonucleases play a crucial role in BER because they recognize the abasic residue and hydrolyze the phosphodiester bond 5′ to the AP site, leaving a gapped DNA intermediate with an extendable 3′-OH end (ref. 2 and references therein).
Members of the family X of DNA polymerases (hereafter, PolX) are widely spread in nature from virus to humans, being involved in the DNA synthesis step during BER and DNA double-strand break repair by virtue of a common Polβ-like core adapted to fill the gapped DNA intermediates very proficiently (6–9).
PolXBs (570-aa long) is a prototypic bacterial/archaeal PolX member from Bacillus subtilis with a N-terminal Polβ-like core (residues 1–317) responsible for catalysis of DNA polymerization (10), and a C-terminal polymerase and histidinol phosphatase (PHP) domain (residues 333–570) containing highly conserved residues that catalyze a Mn2+-dependent 3′–5′-exonuclease activity (11–14), which shows a preferential processing of unannealed 3′ termini (12). Due to this fact and to its adaptation to perform filling of small gaps (10), PolXBs was proposed to play a potential role in the DNA synthesis step of repair pathways during the B. subtilis life cycle, as it has been suggested recently for other bacterial PolXs (10, 15).
Here, we describe the presence of an AP-endonuclease activity intrinsic to PolXBs, genetically linked to the 3′–5′-exonuclease activity and that, in coordination with the polymerization activity, enables the enzyme to recognize, incise, and further repair AP sites. A DNA polymerase with this ability has not been previously reported. The physiological role of PolXBs in a standalone AP-endonuclease-independent DNA repair pathway is discussed.
Results
PolXBs Incises AP Sites.
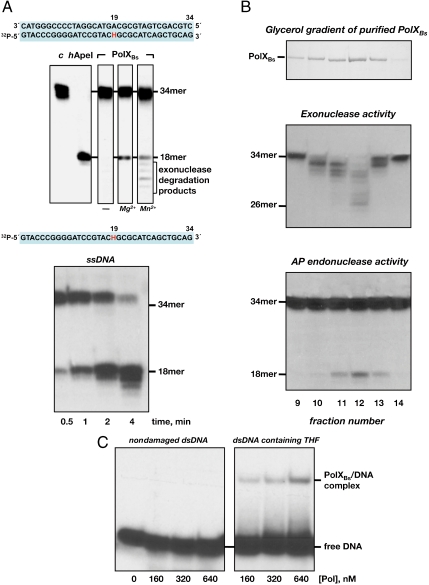
Several PolX members recognize and incise 3′ to abasic sites through a non-metal-dependent β-elimination reaction (AP-lyase activity), giving rise to strand breaks bearing a 3′-phospho-α, β-unsaturated aldehyde (3′-PUA) end that has to be released to allow further elongation (16–18). To ascertain whether PolXBs is endowed with an AP-dependent incision activity, the polymerase was incubated with a 34-mer dsDNA containing an internal tetrahydrofuran (THF), a stable analogue that mimics an abasic site (see Fig. 1A and Materials and Methods). As a control of the possibility of THF to be recognized as an abasic site, this substrate was incubated in parallel with human AP endonuclease I (hApeI), described to break the phosphodiester bond at the 5′ side of THF (19), yielding the expected 18-mer product (Fig. 1A, Upper). As it can be observed, PolXBs did not render any incised product in the absence of metal ions. Conversely, PolXBs possesses a metal-dependent nicking activity on AP sites because Mg2+ and Mn2+ cations promoted the enzyme to cleave at the THF position, giving rise to the 18-mer product. The shorter bands observed with Mn2+ result from the 3′–5′ exonucleolytic degradation of the incised AP site by PolXBs (12), showing that the single-stranded break introduced by the polymerase is prone to further exonucleolysis. These results contrast with the absence of internal cleavage of a nondamaged DNA that is in fact degraded progressively from the 3′ ends by the intrinsic 3′–5′-exonuclease activity of PolXBs, as previously reported (12) (see also Fig. S1). In addition to its activity on dsDNA, PolXBs was also very efficient in introducing an internal nick on ssDNA substrates containing a THF (Fig. 1A, Lower).
Fig. 1.
PolXBs exhibits AP cleavage activity. (A, Upper) Activity on dsDNA. The assay was performed as described in Materials and Methods incubating 1.5 nM of the H/pA dsDNA, 125 nM of PolXBs, and, when indicated, either 8 mM MgCl2 or 1 mM MnCl2 for 30 min at 30 °C. (Lower), activity on ssDNA. The assay was performed as described in Materials and Methods in the presence of 1.5 nM of the [32P] 5′-labeled oligonucleotide H, 125 nM PolXBs, and 1 mM MnCl2. Samples were incubated for the indicated times at 30 °C. H stands for THF. (B) AP cleavage activity is intrinsic to PolXBs. After sedimentation of the purified PolXBs on a 15–30% glycerol gradient (Top), 2 μL of fractions 9–14 were incubated for 1 min at 30°C with 1.5 nM of either the [32P] 5′-labeled pG (Middle) or the [32P] 5′-labeled H (Bottom) oligonucleotide to assay 3′–5′ exonucleolysis and AP nicking activity, respectively, in the presence of 1 mM MnCl2. (C) Binding of PolXBs to dsDNA. The assay was performed as described in Materials and Methods, using as substrate either the pT/pA (nondamaged dsDNA) or the H/pA (dsDNA containing an abasic site), in the presence of the indicated concentration of PolXBs.
To determine whether this activity is inherent to PolXBs, the purified protein was sedimented through a glycerol gradient (Fig. 1B, Top) and the mass peak fractions were assayed for both the 3′–5′ exonuclease (Fig. 1B, Middle) and AP-endonuclease (Fig. 1B, Bottom) activities, using as substrate a 34-mer ssDNA without or with an internal THF site, respectively. Fig. 1B shows that, in both cases, the maximal activity was reached with fraction 12, coincident with the mass peak. The 3′–5′-exonuclease activity exhibited a distributive pattern, as described (12), giving rise to 26–33-mer degradation intermediates. The absence of the 18-mer product rules out the 3′–5′-exonuclease activity as the one responsible for the generation of the product obtained with the THF-containing substrate, confirming the specificity of the nicking activity for an AP site. It is noteworthy that the presence of an AP site precluded the 3′–5′ exonucleolytic degradation from the 3′ end of the THF-containing substrate, as no degradation bands between the 34- and 18-mer products were produced (see Fig. 1B, Bottom). In agreement with the above results, EMSA analysis showed that PolXBs is able to recognize and bind specifically to a dsDNA molecule containing an AP site (Fig. 1C).
PolXBs Possesses an Intrinsic AP-Endonuclease Activity.
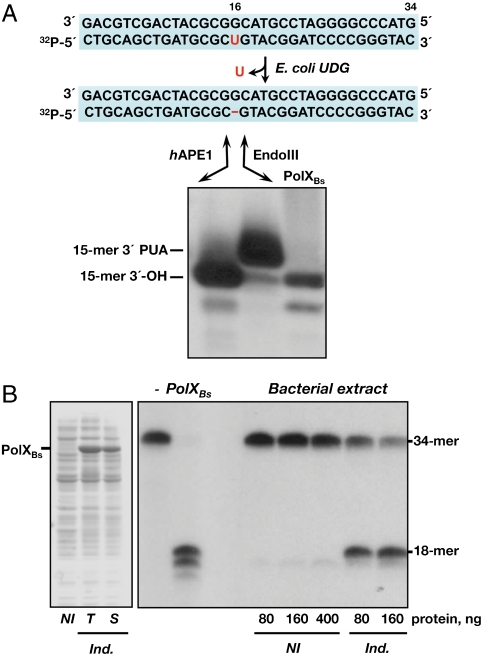
To find out the phosphodiester bond cleaved by PolXBs, the electrophoretic mobility of the nicked strand was compared to that of the products obtained with hApeI and the AP-lyase activity of Escherichia coli endonuclease III (EndoIII). In this case, a uracil containing dsDNA was previously treated with E. coli uracil DNA glycosylase (UDG) to get a natural abasic site because class I glycosylases (as EndoIII) have been reported to be inactive on THF-containing DNA molecules (20). As expected, hApeI hydrolyzed the phosphodiester bond 5′ to the abasic site releasing a 15-mer product with a 3′-OH end (Fig. 2A), whereas EndoIII incised at the 3′ side, leaving a product that migrates slower due to the presence of the resulting blocking PUA moiety at the 3′ end. As observed, the nicked products generated by PolXBs showed identical mobility to those rendered by hApeI, the results being consistent with an AP-endonuclease activity in PolXBs nicking 5′ to the AP site in a metal-dependent manner. The absence of the 15-mer-3′-PUA product agrees with the absence of an AP-lyase activity in PolXBs, in contrast to other PolXs.
Fig. 2.
PolXBs is endowed with AP-endonuclease activity. (A) The uracil-containing dsDNA pU/pG was treated with E. coli UDG, leaving an intact AP site. The resulting AP-containing DNA (1.5 nM) was incubated in the presence of either hApeI that cleaves 5′ to the AP site, EndoIII that incises 3′ to the AP site, or PolXBs. Position of products is indicated. (B) AP-endonuclease activity of E. coli Δxth Δnfo is dependent on the PolXBs expression. (Left) Expression of recombinant PolXBs. (Right) AP-endonuclease activity of bacterial extracts. The experiment was carried out as described in Materials and Methods, incubating 1.5 nM of [32P] 5′-labeled H oligonucleotide with the indicated amount of cellular extract, in the presence of 1 mM MnCl2 at 30 °C for 5 min. NI, noninduced; Ind., induced; T, total extract; S, soluble fraction.
To exclude any possibility of bacterial contamination, PolXBs was expressed in the E. coli strain RPC501 lacking the two endogenous AP endonucleases. As shown in Fig. 2B, incubation of the THF-containing substrate with noninduced bacterial extracts did not give any degradation product, in contrast with those in which PolXBs expression was induced.
Excision of 3′-Blocked Termini by the 3′–5′-Exonuclease Activity of PolXBs.
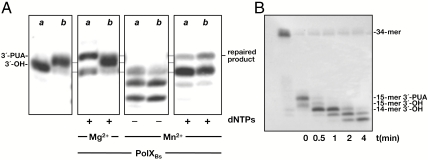
In the course of DNA repair, the action of class I glycosylases, as well as ROS attack on the sugar moiety of DNA, can originate different types of damaged 3′ termini as 3′-PUA, refractory to be elongated by a DNA polymerase. To find out whether PolXBs could restore the 3′-OH end required to prime the gap-filling step during DNA repair of this type of damage, the 15-mer nicked DNA substrates containing 3′-OH and 3′-PUA ends, obtained with hApeI and EndoIII, respectively (see above), were further incubated with PolXBs. As shown in Fig. 3A, in the presence of Mg2+ ions and the four dNTPs PolXBs elongated only the 3′-OH end. The Mn2+-dependent exonuclease activity of PolXBs degraded both 3′ ends, rendering products shorter than 15-mer that were elongated after addition of dNTPs to restore the original nondamaged nucleotide. Time course experiments of 3′–5′ exonuclease on 3′-PUA ends (Fig. 3B) showed a 14-mer as the first degradation product. Thus, contrary to other AP endonucleases that convert the 3′-PUA end to the corresponding 3′-OH form by a 3′-phosphodiesterase activity (3, 21), PolXBs appears to release a dNMP-PUA product by hydrolysis of the penultimate phosphodiester bond. These findings suggest that PolXBs could also play a potential role in cleaning the 3′-blocked ends produced by AP-lyase activities during DNA repair processes. In addition, altogether the results presented above also exclude an AP-lyase activity and further 3′–5′ exonucleolysis of the resulting end as responsible for the generation of the incision products after incubation of an AP-containing substrate with PolXBs.
Fig. 3.
The 3′–5′-exonuclease activity of PolXBs excises 3′-blocked ends. (A) Repair of a 3′-blocked end. The assay was performed as described in Materials and Methods, in the presence of 1.5 nM of either the 18-mer substrate containing 3′-OH (A) or 3′-PUA (B) and 125-nM PolXBs. (B) Exonuclease on a 3′-PUA end. The assay was performed as described in Materials and Methods in the presence of 1.5 nM of the 18-mer substrate containing 3′-PUA end, 125 nM of PolXBs, and 1-mM MnCl2.
DNA Repair of an Abasic Site by PolXBs.
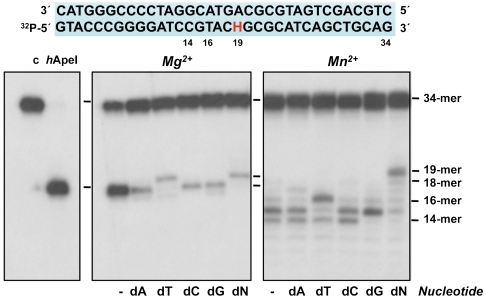
Following the demonstration of an intrinsic AP endonuclease in PolXBs, we evaluated the potential of the polymerase to further extend the resulting 3′ end. As shown in Fig. 4, in the presence of Mg2+ ions, PolXBs added specifically the complementary deoxythymidine monophosphate (dT) opposite the template deoxyadenosine monophosphate (dA) after cleaving 5′ to the THF, giving rise to the repaired 19-mer product and indicating that the AP-endonuclease activity leaves an extendable 3′-OH end. Even in the presence of the four nucleotides, the polymerase catalyzed solely the addition of one nucleotide, displacing the dangling downstream 5′-THF moiety. As expected, Mn2+ ions promoted the exonucleolytic degradation of the cleaved strand, enlarging the nick into a gap, yielding 14–16-mer products that were further extended by addition of the nucleotide complementary to the first template position. Thus, the 14-mer product was extended specifically with deoxyguanosine monophosphate, the 15-mer with dT, and the 16-mer with dA. Addition of the four nucleotides made PolXBs to refill the gap to completion, the last dT getting access to the position initially occupied by THF, restoring the original nucleotide. Hence, PolXBs is able to reallocate the resulting 3′-OH primer terminus at the polymerization active site to restore (repair) the original nucleotide.
Fig. 4.
PolXBs restores the original nucleotide in the absence of standalone AP endonucleases. The assay was performed as described in Materials and Methods incubating 1.5 nM of the H/pA dsDNA, 125 nM PolXBs, either 8 mM MgCl2 or 1 mM MnCl2, and 100 μM of the indicated nucleotide for 30 min at 30 °C.
AP-Endonuclease Activity of PolXBs Maps at the PHP Domain.
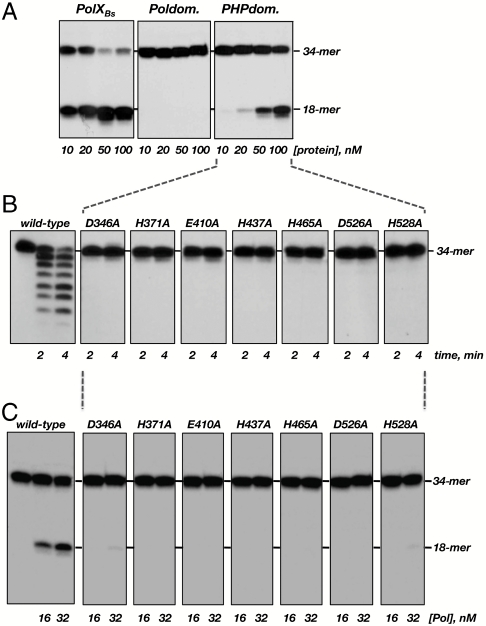
To map the PolXBs domain responsible for the AP-endonuclease activity, the presence of the latter was analyzed in the separately expressed and purified polymerization and PHP domains (12). Fig. 5A shows that the AP-endonuclease activity is specifically present in the PHP domain. The 10-fold reduction of the catalytic efficiency exhibited by this domain respect to the complete enzyme suggests that proficient endonucleolysis would require the assistance of the polymerization domain.
Fig. 5.
AP-endonuclease and 3′–5′-exonuclease activities are genetically linked. (A) AP-endonuclease activity of PolXBs is located at the C-terminal PHP domain. The assay was carried out in the conditions described in Materials and Methods, incubating the indicated concentration of either the complete PolXBs or the independent Polymerization (Poldom) and PHP (PHPdom) domains with 1.5 nM of the [32P] 5′-labeled oligonucleotide H and 1-mM MnCl2 for 1 min at 30 C. (B) Exonuclease activity of mutants in metal ligands of the PHP domain. The assay was performed incubating 125 nM of either the wild-type or mutant polymerase, 1.5 nM of [32P] 5′-labeled oligonucleotide pG, and 1-mM MnCl2 for the indicated times at 30 °C. (C) AP-endonuclease activity of mutants in metal ligands of the PHP domain. The assay was carried out incubating the indicated amount of either the wild-type or mutant polymerase, 1.5 nM of the ssDNA containing THF, and 1-mM MnCl2 for 30 s at 30 °C.
Multiple sequence alignment of the PHP domain of bacterial/archaeal PolXs permitted the identification of highly conserved residues grouped in four motifs, assembling a catalytic core that would coordinate the metal ions responsible for the 3′–5′-exonuclease activity (12). To ascertain the involvement of the PHP active site in supporting catalysis of both, the 3′–5′-exonuclease and AP-endonuclease activities, mutants D346A (motif I), H371A (motif II), E410A and H437A (motif III), H465A, and D526A and H528A (motif IV), at the corresponding PolXBs residues, were obtained (see Materials and Methods). As shown in Fig. 5 B and C, the changes introduced severely impaired the two enzymatic activities, indicating that they are genetically linked and share the same catalytic active site. The polymerization activity of the mutant derivatives respect to the wild-type enzyme ranged from 50% to 100% (see Table S1), excluding a global misfolding as the cause of the specific lack of AP-endonuclease and 3′–5′-exonuclease activities.
Discussion
Intracellular ROS and exogenous genotoxic agents damage DNA, leading directly or indirectly to the formation of AP sites and strand breaks. In most organisms, AP endonucleases, essential components of the BER pathway, recognize and promote repair of these DNA lesions (reviewed in refs. 2 and 3).
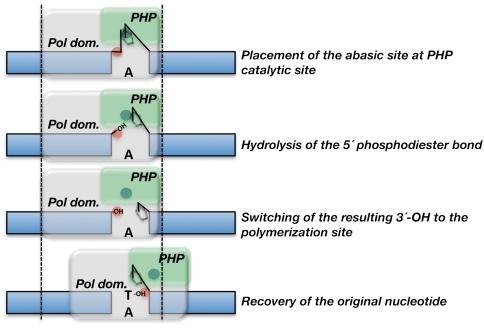
In this paper, we have shown that PolXBs is endowed with an intrinsic AP-endonuclease activity that cleaves 5′ to an abasic site in a metal-dependent manner. Functional coordination of AP-endonuclease and polymerization activities enables the polymerase to recognize, incise, and further restore in vitro the genetic information of the damaged DNA back to its original state in the absence of additional factors (see scheme in Fig. 6). The biochemical analysis of the independent PolXBs domains has demonstrated the location of the AP-endonuclease activity at the C-terminal PHP domain, being the previously undescribed presence of an intrinsic AP-endonuclease activity in a member of the family X of DNA polymerases.
Fig. 6.
Scheme depicting the proposed steps during the repair of AP sites by PolXBs. Polymerization and PHP domains are represented as gray and green boxes, respectively. Polymerization and 3′–5′-exonuclease/AP-endonuclease active sites are shown as red and dark green circles, respectively.
The PHP domain constitutes a family of phosphoesterases associated with the N terminus of the α subunit of bacterial DNA polymerase III and bacterial/archaeal PolXs (11). This domain shows an unusual topology of α7β7 barrel with a cleft at its C-terminal side where invariant His, Asp, and Glu residues are involved in coordination of three metal ions (11, 13, 22–24). Analysis of mutants at the corresponding residues of the PHP domain of PolXBs has shown the involvement of these metal ligands in the catalysis of the 3′–5′ exo- and AP endonucleolysis, both activities being genetically linked. Despite the lack of amino acid similarities, the prototypic E. coli endonuclease IV (EndoIV) is structurally similar to the PHP domains, showing a triosephosphate-isomerase-barrel topology. The three Zn2+/Mn2+ ions that catalyze the phosphodiester bond hydrolysis are also coordinated by conserved His, Asp, and Glu residues arranged like the PHP metal ligands (24–26) (see Fig. S2A and SI Discussion). In addition, besides its AP endonuclease, EndoIV also possesses a 3′–5′-exonuclease activity governed by the same active site (25, 27–29). Altogether, the results lead us to propose for the PHP domain of bacterial/archaeal PolXs a three metal ions mechanism similar to EndoIV for recognition of damaged DNA, binding, and incision, suggesting a convergent evolution to give rise to a 3′-OH end required to prime further DNA repair synthesis. The higher affinity exhibited by PolXBs for DNA substrates containing AP sites with respect to nondamaged DNA suggests that the polymerase could bind DNA and further scan it in search for abasic sites, as suggested for other AP endonucleases (26).
Many organisms have a duplication of AP endonucleases, most probably as a result from the critical nature of the BER pathway for cell survival. Thus, whereas organisms as E.coli and Saccharomyces cerevisiae express ExoIII and EndoIV, representatives of the structural distinct families Xth and Nfo AP endonucleases (30, 31), others as Neisseria meningitidis and human cells contain two AP endonucleases belonging to the Xth-type family (32, 33). B. subtilis possesses two known AP endonucleases (34), ExoA (Xth), which is expressed in growing cells and in the forespore compartment of the sporulating cell (34), and Nfo, present only in dormant spores (35). Thus, based on their differential expression pattern, only ExoA would be present during the vegetative growth of cell. The fact that loss of both proteins neither decreased resistance to oxidative agents nor increased the spontaneous mutation frequency in growing cells (35) opened the possibility of the existence of a third unknown AP-endonuclease activity in B. subtilis to generate extendable 3′-OH ends during BER. Thus, PolXBs may act as a backup mechanism providing the additional AP-endonuclease activity, taking part of a potentially new DNA repair pathway, as it contains three of the activities that are usually catalyzed by standalone proteins in BER: one is the AP endonuclease to break the phosphodiester bond 5′ to the AP site; another is the 3′–5′ exonuclease to release potential 3′-blocking groups arisen by an AP-lyase activity, as that of the bifunctional class I glycosylases MutM or Nth that release oxidized base lesions. Conservation of a 3′–5′ exonuclease among the major AP endonucleases emphasizes the significance of this activity in a damage-cleansing function (28, 29, 36). As described in Results, the presence of an abasic site in a linear DNA hinders exonucleolytic degradation of the 3′ ends. This fact together with the fact that both activities depend on the same active site could indicate that preferential binding to the AP residue prevents binding to the 3′ end, guaranteeing a sequential action of the AP endonuclease and further exonucleolysis. The result of these two activities is the generation of a 3′-OH end that will be further elongated by the third activity, DNA polymerization, responsible for the gap-filling step to restore (repair) the original nucleotide. Our findings support the current thought that the presence of multiple versions of DNA repair enzymatic activities in a single organism reflects their fundamental importance for cellular viability (32).
As it has been shown in this work, PolXBs AP endonuclease can also hydrolyze AP sites in ssDNA. This property has been reported in AP endonucleases as hApeI (37) and Chlamydia pneumoniae AP endonuclease IV (38). In addition, other enzymes involved in BER, as several phylogenetically diverse DNA N-glycosylases, have shown activity against damaged bases in ssDNA (39–45). The presence of abasic sites in ssDNA regions is highly deleterious because, in addition to block replication and transcription, the nicking action of a canonical AP endonuclease could induce lethal dsDNA breaks. Based on the findings described here, a role for PolXBs to repair these lesions on ssDNA could be postulated, likely acting in concert with additional protein factors to prevent separation of the two ssDNA regions originated after the action of the AP-endonuclease activity, as it has been proposed to occur in other systems (37).
Materials and Methods
Enzymes and Reagents.
Unlabeled nucleotides were purchased from GE Healthcare. [γ-32P]ATP (3,000 Ci/mmol) and [α-32P]dATP (3,000 Ci/mmol) were obtained from Perkin Elmer. T4 polynucleotide kinase (T4PNK), hApeI, E. coli UDG, and EndoIII were obtained from New England Biolabs. PolXBs, Poldom, and PHPdom deletion mutants were expressed and purified as described (10, 12). The purified PolXBs was further loaded into a 5 mL glycerol gradient (15–30%) containing 50 mM Tris•HCl, pH 7.5, 0.2 M NaCl, 1 mM EDTA, and 7 mM β-mercaptoethanol, and centrifuged at 62,000 rpm (Beckman SW.50 rotor) for 26 h at 4 °C. After centrifugation, 20 fractions were collected from the bottom of the tube.
Oligonucleotides, DNA Templates and Substrates.
Oligonucleotides H (5′-GTACCCGGGGATCCGTACHGCGCATCAGCTGCAG), where H stands for THF, pT (5′-GTACCCGGGGATCCGTACTGCGCATCAGCTGC), and pU (5′-CTGCAGCTGATGCGCUGTACGGATCCCCGGGTAC) were 5′ labeled with [γ-32P]ATP and T4PNK and hybridized to pA (5′CTGCAGCTGATGCGCAGTACGGATCCCCGGGTAC) or pG (5′-GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG), as indicated, to form dsDNA substrates. All the hybridizations were performed in the presence of 0.2 M NaCl and 60 mM Tris•HCl, pH 7.5.
AP-Endonuclease Activity Assays.
The incubation mixtures contained, in 12.5 μL, 50 mM Tris•HCl, pH 7.5, either 1 mM MnCl2 or 8 mM MgCl2, 1 mM DTT, 4% glycerol, 0.1 mg/mL BSA, 125 nM of the wild-type or mutant PolXBs or 2 μL of the different fractions from a glycerol gradient. As substrate, either 1.5 nM of 5′ labeled oligonucleotide H (ssDNA) or duplex H/pA was used. When indicated, 100 μM of the specified nucleotide was also added. Samples were incubated at 30 °C for the indicated times and quenched by adding 10 mM EDTA. Reactions were analyzed by 8 M urea-20% PAGE and autoradiography.
AP-Endonuclease Activity Assay on Uracil Containing 5′ labeled substrates.
The 34-mer oligonucleotide pU was 5′ labeled and hybridized to pG. This dsDNA (3 nM) was treated with 2 units of E. coli UDG for 10 min at 37 °C in the presence of 50 mM Tris•HCl, pH 7.5, 1 mM DTT, 4% glycerol, and 0.1 mg/mL BSA. After incubation, the mixture was supplemented with 1 mM MnCl2 and treated with either 10 units hAPE1, 10 units of EndoIII, or 125 nM of PolXBs. Reactions were incubated for 30 min at 37 °C and quenched by adding 10 mM EDTA. Reactions were analyzed by 8-M urea-20% PAGE and autoradiography.
Electrophoretic Mobility Shift Assay.
The incubation mixture contained, in a final volume of 20 μL, 50 mM Tris•HCl, pH 7.5, 1 mM MnCl2, 1 mM DTT, 4% glycerol, 0.1 mg/mL BSA, 0.7 nM of either H/pA or pT/pA dsDNA, and the indicated amount of PolXBs. After incubation for 20 min at 30 °C, the samples were subjected to electrophoresis in precooled 4% (wt/vol) polyacrylamide gels (80∶1, monomer∶bis) containing 12 mM Tris-acetate (pH 7.5) and 1 mM EDTA, and run at 4 °C in the same buffer at 8 V/cm (46).
Exonuclease Activity on 5′ Labeled DNA Substrates.
The incubation mixture contained, in 12.5 μL, 50 mM Tris•HCl, pH 7.5, 1 mM MnCl2, 1 mM DTT, 4% glycerol, 0.1 mg/mL BSA, and 125 nM of wild-type or mutant PolXBs. As substrate, 1.5 nM of 5′ labeled pG was used. Samples were incubated at 30 °C for 1 min and quenched by adding 10 mM EDTA. Reactions were analyzed by 8 M urea-20% PAGE and autoradiography. DNA (1.25 nM) containing either 3′-OH or 3′-PUA ends, obtained as described above, was used as substrate of the exonuclease activity in the presence of either 8 mM MgCl2 or 1 mM MnCl2, for either 1 min or the indicated times at 30 °C. When indicated, 100 μM dNTPs were added to the reaction.
Site-Directed Mutagenesis of PolXBs.
PolXBs mutants D346A, H371A, E410A, H437A, H465A, D526A, and H528 were obtained by using the QuickChange site-directed mutagenesis kit obtained from Amersham Pharmacia. Plasmid pET28-PolXBs containing the PolXBs gene was used as template for the reaction. Expression and purification of the mutant proteins were performed as described for the wild-type PolXBs (10).
Expression of PolXBs in E. coli Strain RPC501.
B. subtilis yshc gene was cloned into pT7-3 expression vector under the control of the T7 RNA polymerase-specific ϕ10 promoter (47). E. coli AP-endonuclease genes xth and nfo are deleted by introduction of Chlr and Kanr genes, respectively, in strain RPC501 (48). This host was further lysogenized by site-specific integration of λDE3 prophage into the bacterial chromosome by using the λDE3 Lysogenization Kit following the manufacturer protocol (Novagen). The lysogenized host was transformed with plasmid pT7-3-PolXBs, and PolXBs expression was induced with 0.5-mM IPTG. Cell extracts were prepared by sonication of a liquid culture of both noninduced and induced cells (OD = 0.6) and further centrifugation (14,000 rpm at 4 °C during 15 min in a Hettich Zentrifugen Mikro 22R) to get the soluble fraction. Protein amount in soluble extracts was quantitated by Bradford.
DNA Polymerization Activity Assays on Activated DNA.
The assay was performed essentially as described (10). Polymerization activity of mutant derivatives with respect to that of the wild-type enzyme (percentage) was calculated from the Cerenkov radiation of the excluded volume.
Supplementary Material
Acknowledgments.
We are grateful to Richard P. Cunningham (Albany, NY) who kindly provided E. coli strain RPC501, and to José M. Lázaro for advice in protein purification. This work was supported by the Spanish Ministry of Science and Innovation Grants Consolider-Ingenio CSD2007-00015 and BFU2008-00215 (to M.S.), by Spanish Research Council Grant 200920I012 (to M.d.V.), and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” B.B. is a recipient of a fellowship from Fundación Ramón Areces.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013603107/-/DCSupplemental.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Krwawicz J, Arczewska KD, Speina E, Maciejewska A, Grzesiuk E. Bacterial DNA repair genes and their eukaryotic homologues: 1. Mutations in genes involved in base excision repair (BER) and DNA-end processors and their implication in mutagenesis and human disease. Acta Biochim Pol. 2007;54:413–434. [PubMed] [Google Scholar]
- 3.Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005;33:4404–4411. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braithwaite EK, et al. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 7.García-Díaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: A possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava DK, et al. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 10.Baños B, Lázaro JM, Villar L, Salas M, de Vega M. Characterization of a Bacillus subtilis 64-kDa DNA polymerase X potentially involved in DNA repair. J Mol Biol. 2008;384:1019–1028. doi: 10.1016/j.jmb.2008.09.081. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Koonin EV. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baños B, Lázaro JM, Villar L, Salas M, de Vega M. Editing of misaligned 3′-termini by an intrinsic 3′–5′ exonuclease activity residing in the PHP domain of a family X DNA polymerase. Nucleic Acids Res. 2008;36:5736–5749. doi: 10.1093/nar/gkn526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leulliot N, et al. The family X DNA polymerase from Deinococcus radiodurans adopts a non-standard extended conformation. J Biol Chem. 2009;284:11992–11999. doi: 10.1074/jbc.M809342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakane S, Nakagawa N, Kuramitsu S, Masui R. Characterization of DNA polymerase X from Thermus thermophilus HB8 reveals the POLXc and PHP domains are both required for 3′–5' exonuclease activity. Nucleic Acids Res. 2009;37:2037–2052. doi: 10.1093/nar/gkp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khairnar NP, Misra HS. DNA polymerase X from Deinococcus radiodurans implicated in bacterial tolerance to DNA damage is characterized as a short patch base excision repair polymerase. Microbiology. 2009;155:3005–3014. doi: 10.1099/mic.0.029223-0. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Escudero R, Garcia-Diaz M, Salas ML, Blanco L, Salas J. DNA polymerase X of African swine fever virus: Insertion fidelity on gapped DNA substrates and AP lyase activity support a role in base excision repair of viral DNA. J Mol Biol. 2003;326:1403–1412. doi: 10.1016/s0022-2836(03)00019-6. [DOI] [PubMed] [Google Scholar]
- 17.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 18.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 19.Demple B, Sung J-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair. 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 21.Vidal AE, et al. Crystal structure and DNA repair activities of the AP endonuclease from Leishmania major. J Mol Biol. 2007;373:827–838. doi: 10.1016/j.jmb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 23.Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 24.Teplyakov A, et al. Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site. Proteins. 2003;51:315–318. doi: 10.1002/prot.10352. [DOI] [PubMed] [Google Scholar]
- 25.Garcin ED, et al. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol. 2008;15:515–522. doi: 10.1038/nsmb.1414. [DOI] [PubMed] [Google Scholar]
- 26.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 27.Golan G, Ishchenko AA, Khassenov B, Shoham G, Saparbaev MK. Coupling of the nucleotide incision and 3′ → 5′ exonuclease activities in Escherichia coli endonuclease IV: Structural and genetic evidences. Mutat Res. 2009;685:70–79. doi: 10.1016/j.mrfmmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Ishchenko AA, Yang X, Ramotar D, Saparbaev M. The 3′ → 5′ exonuclease of Apn1 provides an alternative pathway to repair 7, 8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerins SM, Collins R, McCarthy TV. Characterization of an endonuclease IV 3′–5′ exonuclease activity. J Biol Chem. 2003;278:3048–3054. doi: 10.1074/jbc.M210750200. [DOI] [PubMed] [Google Scholar]
- 30.Boiteux S, Guillet M. Abasic sites in DNA: Repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Ljungquist S, Lindahl T, Howard-Flanders P. Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol. 1976;126:646–653. doi: 10.1128/jb.126.2.646-653.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter EP, et al. AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. EMBO J. 2007;26:1363–1372. doi: 10.1038/sj.emboj.7601593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hadi MZ, Wilson DM., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36:312–324. [PubMed] [Google Scholar]
- 34.Salas-Pacheco JM, Urtiz-Estrada N, Martínez-Cadena G, Yasbin RE, Pedraza-Reyes M. YqfS from Bacillus subtilis is a spore protein and a new functional member of the type IV apurinic/apyrimidinic-endonuclease family. J Bacteriol. 2003;185:5380–5390. doi: 10.1128/JB.185.18.5380-5390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salas-Pacheco JM, Setlow B, Setlow P, Pedraza-Reyes M. Role of the Nfo (YqfS) and ExoA apurinic/apyrimidinic endonucleases in protecting Bacillus subtilis spores from DNA damage. J Bacteriol. 2005;187:7374–7381. doi: 10.1128/JB.187.21.7374-7381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanno S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marenstein DR, Wilson DM, 3rd, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair. 2004;3:527–533. doi: 10.1016/j.dnarep.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Liu J. Chlamydia pneumoniae AP endonuclease IV could cleave AP sites of double- and single-stranded DNA. Biochim Biophys Acta. 2005;1753:217–225. doi: 10.1016/j.bbapap.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Boorstein RJ, et al. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 40.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 41.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishchenko AA, Bulychev NV, Maksakova GA, Johnson F, Nevinsky GA. Single-stranded oligodeoxyribonucleotides are substrates of Fpg protein from Escherichia coli. IUBMB Life. 1999;48:613–618. doi: 10.1080/713803570. [DOI] [PubMed] [Google Scholar]
- 43.Kavli B, et al. hUNG2 is the major repair enzyme for removal of uracil from U∶A matches, U∶G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 44.Kumar NV, Varshney U. Contrasting effects of single stranded DNA binding protein on the activity of uracil DNA glycosylase from Escherichia coli towards different DNA substrates. Nucleic Acids Res. 1997;25:2336–2343. doi: 10.1093/nar/25.12.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takao M, et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 46.Carthew RW, Chodosh LA, Sharp PA. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- 47.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.