Abstract
DNA glycosylases initiate the base excision repair (BER) pathway by excising damaged, mismatched, or otherwise modified bases. Animals and plants independently evolved active BER-dependent DNA demethylation mechanisms important for epigenetic reprogramming. One such DNA demethylation mechanism is uniquely initiated in plants by DEMETER (DME)-class DNA glycosylases. Arabidopsis DME family glycosylases contain a conserved helix–hairpin–helix domain present in both prokaryotic and eukaryotic DNA glycosylases as well as two domains A and B of unknown function that are unique to this family. Here, we employed a mutagenesis approach to screen for DME residues critical for DNA glycosylase activity. This analysis revealed that amino acids clustered in all three domains, but not in the intervening variable regions, are required for in vitro 5-methylcytosine excision activity. Amino acids in domain A were found to be required for nonspecific DNA binding, a prerequisite for 5-methylcytosine excision. In addition, mutational analysis confirmed the importance of the iron-sulfur cluster motif to base excision activity. Thus, the DME DNA glycosylase has a unique structure composed of three essential domains that all function in 5-methylcytosine excision.
Keywords: DNA repair, helix–hairpin–helix protein, mutagenesis, mixed charge cluster
The modified base 5-methylcytosine (5mC) is a stable epigenetic mark that silences gene expression and plays an important role in many developmental processes such as gene imprinting, X-chromosome inactivation, and transposon silencing (1–4). DNA methylation primarily occurs at symmetric CG sequences in animals, whereas DNA methylation in plants occurs in all sequence contexts: CG, CHG, and CHH (where H = A, T, or C) (5). The overall CG DNA methylation pattern in the genome is faithfully inherited to daughter cells by maintenance DNA methyltransferases, which convert hemimethylated sites generated by DNA replication to fully methylated sites. When maintenance methyltransferases are absent or down-regulated, DNA methylation is progressively lost during replication, which is referred to as passive DNA demethylation. By contrast, active DNA demethylation that is independent of DNA replication requires alternative pathways.
Base excision repair (BER), which normally functions to repair damaged and mispaired bases, is also required for active DNA demethylation and epigenetic reprogramming in eukaryotes (3, 6–9). BER is initiated by DNA glycosylase enzymes that catalyze the hydrolysis of the N-glycosidic (base-ribose) bond. In plants, the DEMETER (DME) family of DNA glycosylases functions to remove 5mC, which is then replaced by unmethylated cytosine (10, 11), resulting in transcriptional activation of target genes (10, 12). Arabidopsis has three other DME-like (DML) genes—ROS1, DML2, and DML3 (12–14) (Fig. S1). DME is essential for plant reproduction and influences the endosperm DNA methylation profile (15, 16), whereas DMLs function in vegetative tissues to prevent inappropriate gene silencing and maintain the genome-wide methylation profiles (13, 14, 17, 18). Previously we showed that DME is a bifunctional DNA glycosylase/AP-lyase that excises 5mC from double-stranded DNA regardless of the sequence context to produce β- and δ-elimination products in conjunction with cleavage of the phosphodiester bond (10). ROS1 also excises 5mC (11, 14), and a recent study proposed that 5mC excision by ROS1 occurs in a distributive manner on long DNA substrates (19).
DME family DNA glycosylases have both common and unique structural and functional features compared to typical DNA glycosylases. The glycosylase domain of DME contains a helix–hairpin–helix (HhH) motif and a glycine/proline-rich loop with a conserved aspartic acid (GPD), also found in human 8-oxoguanine DNA glycosylase (hOGG1), Escherichia coli adenine DNA glycosylase (MutY), and endonuclease III (Endo III) (20–22). In addition, DME possesses four cysteine residues adjacent to the DNA glycosylase domain that may function to hold a [4Fe-4S] cluster in place as in MutY and Endo III (21, 22). In contrast to most other members of the HhH glycosylase superfamily, DME family members contain two additional conserved domains (domain A and domain B) flanking the central glycosylase domain (Fig. S1).
The unique structure and function of DME family DNA glycosylases raises questions regarding the roles of the individual domains. Is the DME glycosylase domain, which shares some common features with other HhH-[4Fe-4S] glycosylases, functionally conserved? What is the function of the conserved A and B domains that are unique to the DME-class DNA glycosylases? To address these questions, we carried out extensive mutagenesis of DME to identify the domains and amino acid residues required for in vitro excision of 5mC. We found that the glycosylase domain of DME retains essential features of the HhH-GPD superfamily and that the [4Fe-4S] cluster motif is required for 5mC excision. Mutation profiles reveal that DME has a modular structure consisting of three conserved domains required for biochemical activity. We also report that DNA binding activity of domain A is a prerequisite for 5mC excision in vitro. This study provides previously undescribed insight into the unique structure and function of 5mC glycosylases necessary for DNA demethylation.
Results
DME Requires All Three Conserved Domains for DNA Demethylation Activity.
Previously we showed that DME lacking the N-terminal 537 amino acids (DMEΔN537) contains glycosylase activity at a 5mC residue in an oligonucleotide substrate (10). In an effort to identify the minimal regions for glycosylase activity, a series of deletions were made on both N- and C-terminal ends of DME. We produced DME fragments lacking N-terminal 677 (DMEΔN677) and 896 (DMEΔN896) amino acids, respectively. DMEΔN677, like DMEΔN537, displayed 5mC glycosylase activity (Fig. S1). By contrast, we did not detect activity in reactions with DMEΔN896 (Fig. S1). A small deletion at the C terminus (51 amino acids) was enough to completely abolish the glycosylase activity when combined with the otherwise active DMEΔN677 (Fig. S1). All active fragments tested retain the three conserved domains for 5mC glycosylase activity (Fig. S1).
Iron–Sulfur Cluster Motif Is Necessary for DME Activity.
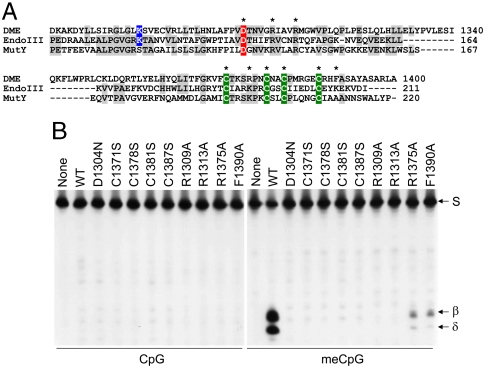
In the HhH superfamily, several DNA glycosylases such as Endo III and MutY possess the [4Fe-4S] cluster. Despite a significant homology with them, however, E. coli 3-methyladenine DNA glycosylase (AlkA) and hOGG1 lack the [4Fe-4S] cluster (20, 23, 24), which led us to investigate whether the same type of cluster predicted in DME is required for the glycosylase activity. DME has four cysteine residues with the spacing characteristic of the HhH-[4Fe-4S] glycosylase superfamily: Cys1371-X6-Cys1378-X2-Cys1381-X5-Cys1387 (Fig. 1A). All cysteine mutant DME proteins showed no 5mC excision activity in vitro (Fig. 1B). These findings suggest that four cysteines in the [4Fe-4S] cluster motif of DME are necessary for catalytic activity and/or stability of DME.
Fig. 1.
Importance of the [4Fe-4S] cluster of DME for 5mC excision. (A) A HhH-GPD/Fe-S cluster of DME is structurally similar to those of EndoIII and MutY in E. coli. Besides a conserved HhH motif, four cysteine residues (green) that constitute the [4Fe-4S] cluster and some other functionally important residues are highly conserved among these three proteins. Catalytically important aspartic acid (canonical in this family) and lysine (specific to bifunctional enzymes with glycosylase/AP-lyase activities) residues are colored in red and blue, respectively. The amino acid residues subjected to site-directed mutagenesis are indicated with asterisks. (B) In vitro 5mC excision activity of [4Fe-4S] cluster mutant DME proteins. Active MBP-DMEΔN677 (WT) and proteins with indicated amino acid substitutions were reacted with unmethylated (Left) or methylated (Right) oligonucleotide substrates. Oligonucleotide substrate (S) and β- and δ-elimination products are indicated to the right of the panel.
To further investigate the nature of the [4Fe-4S] cluster motif of DME, several conserved residues presumably associated with the structure and/or function of the cluster were chosen and subjected to site-directed mutagenesis. A protruding loop formed by the first two cysteine residues in the [4Fe-4S] cluster, termed the iron–sulfur cluster loop (FCL) motif, contains a high density of positively charged residues in Endo III and MutY (Fig. 1A) that are properly positioned to interact with a negatively charged DNA backbone (21, 25). In addition, two positive arginine residues adjacent to the invariant aspartic acid (Asp-X4-Arg-X3-Arg) are known to form a hydrogen bond with the FCL loop in Endo III and MutY (25–27) (Fig. 1A). In order to confirm whether such corresponding residues in DME are essential for enzyme activity, as in other [4Fe-4S] glycosylases, Arg1309, Arg1313, or Arg1375 of DME was substituted with alanine. We did not detect glycosylase activity in reactions with DMEΔN677 with an R1309A or R1313A substitution, and DMEΔN677(R1375A) displayed a significant decrease in base excision activity (Fig. 1B). Another conserved element found in the vicinity of the [4Fe-4S] cluster is a large hydrophobic residue, which has been proposed to protect the cluster from water (27). DME has several “bulky” residues (Phe1390 and Tyr1394) immediately downstream of the [4Fe-4S] cluster motif, reminiscent of Trp216 in MutY (Fig. 1A). We found that DMEΔN677 (F1390A) displayed a significant decrease in glycosylase activity (Fig. 1B). Taken together, these findings suggest that four cysteines and other residues may comprise an [4Fe-4S] cluster that is essential for DME glycosylase activity.
Random Substitutions Reveal Essential Residues for DME Activity.
The HhH glycosylase domain is the core-conserved element of this class of DNA glycosylases (28). Previously, we showed that two catalytically important residues (K1286 and D1304) in the HhH motif are necessary for the bifunctional glycosylase/AP-lyase activity of DME (10). However, little is known about the contribution of other amino acids to catalytic activity, base recognition, and/or folding. Previously, Guo et al. (29) randomly mutated 3-methyladenine DNA glycosylase and identified amino acid substitutions that did not affect enzyme activity. In contrast to their approach to assess protein “tolerance” to random amino acid change, we investigated protein “susceptibility” to random amino acid changes by revealing the substitutions critical for DME activity. To identify functionally essential residues of DME that reside throughout the protein, we randomly mutated and identified mutant DME proteins that lacked enzyme activity by expressing them in a bacterial system. As a bifunctional DNA glycosylase, DME is thought to produce abasic sites and single-strand breaks in the course of base excision. These lesions are detrimental to DNA replication and transcription in bacteria unless immediately repaired. In accordance to this idea, the expression of DME affects viability of methylation-proficient E. coli strains (10), due probably to excessive accumulation of such harmful lesions in the bacterial genome.
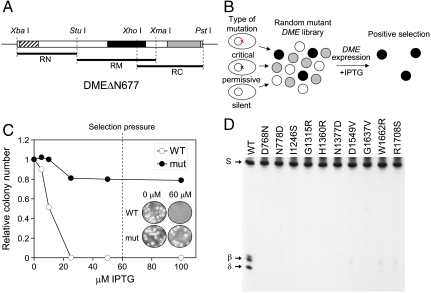
Taking advantage of the lethal effect of DME expression on E. coli, we developed a screening strategy to identify functionally essential residues for the glycosylase activity of DME (Fig. 2 A and B). First, the coding region of active DMEΔN677 was divided into three regions (RN, RM, and RC represent the N-terminal, mid, and C-terminal regions of DMEΔN677, respectively; Fig. 2A), and each was subjected to random mutagenesis. Mutagenized fragments were cloned in an expression vector replacing the wild-type sequence. Subsequently, the constructs were transformed into the methylation-proficient E. coli strain DH5α, generating a random mutant library. Transformed cells were grown on IPTG medium to induce DME expression and surviving colonies were isolated. E. coli cells that expressed a DME sequence in which a mutation(s) abolished activity were predicted to survive and form colonies, as harmful base excision would not occur. By contrast, cells expressing active DME with no mutations, or a silent or permissive mutation(s), would suffer excessive base excision and would not survive.
Fig. 2.
Isolation of mutant DME proteins sensitive to random amino acid change. (A) DMEΔN677 was divided into three pieces (RN, RM, and RC) and subjected to random mutagenesis by error-prone PCR. Three conserved domains—domain A, glycosylase domain, and domain B—are indicated by hatched, solid, and shaded boxes, respectively. The restriction sites used for cloning are indicated above the DME fragment. (B) Screening scheme of a random mutant library. A mutant library was generated and screened for viability in the presence of IPTG to induce DME expression. Only cells with a critical mutation(s) will survive due to a loss of 5mC glycosylase activity that would otherwise produce critical damages in E. coli. Surviving colonies were positively selected and analyzed for enzyme activity. (C) The cells expressing catalytically inactive DME survived, whereas the cells expressing WT DME died as IPTG concentrations increased. A dashed line represents the concentration of IPTG (60 μM) in media that exerted selection pressure. (Inset) Colony formation of wild-type and mutant (D1304N) DME transformants at 0 and 60 μM IPTG. (D) 5mC glycosylase activity of some representative mutant DME proteins. WT and mutant proteins were reacted with methylated oligonucleotide substrate and separated on a denaturing polyacrylamide gel.
To test the system, we induced expression of active DMEΔN677 with IPTG and verified that the colony formation significantly decreased as the IPTG concentration on the medium increased (Fig. 2C). By contrast, IPTG-induced expression of catalytically inactive DMEΔN677 (K1286Q) had little effect on cell viability. Plasmids were isolated from the surviving colonies, and mutations were detected by determining their DNA sequence. As expected, all isolated plasmids contained one or multiple mutations. Here, we report results when only a single amino acid was changed so that function could be unambiguously assigned. We isolated a total of 102 clones with single amino acid changes that decreased the base excision activity of DME in E. coli (no cytotoxicity), and 85 of them were unique substitutions at 75 positions (Table S1). Mutations were distributed over the entire DMEΔN677 sequence (Table S1 and Fig. S2) and displayed all possible base transitions.
Verifying the reliability of the screening, some substitutions occurred at functionally important residues, whose requirement had already been confirmed by site-directed mutagenesis (Fig. 1B). For instance, three mutant clones RM7-38, RM7-54, and RM7-115 harbored a substitution of a conserved catalytic residue Lys at position 1286 to Asn, Arg, and Glu, respectively (Table S1 and Fig. S3). Two substitutions were also found in the cysteine residues in the [4Fe-4S] cluster motif—clones RM2-30 and RC1-142 had C1381R and C1387R substitutions, respectively.
Consistent with the loss of in vivo activity (no cytotoxicity) in E. coli, all the mutant proteins were unable to excise 5mC from methylated DNA in vitro. Some representative reactions are shown in Fig. 2D (for additional reactions, see Fig. S4). We therefore conclude that the individual amino acid residues where substitutions have occurred are crucial for in vitro activity of DME and that every single substitution had a negative effect on 5mC glycosylase activity and/or stability.
Three Distinct Domains Are Necessary and Sufficient for 5mC Excision.
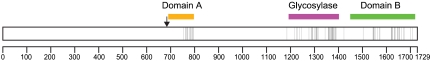
Interestingly, when all the site-directed and random substitutions were mapped on the coding region of DME, their distribution was clustered rather than random (Fig. 3). The majority of substitutions were confined to the three conserved domains, with a gap between domain A and the glycosylase domain, where no substitutions were identified. Also, many of the amino acid substitutions are associated with predicted α-helices and β-sheets (Fig. S2). These findings demonstrate that the three conserved domains with a high density of secondary structures are functionally important and more susceptible to a subtle structural change caused by an amino acid substitution. By contrast, nonconserved regions appeared to be more tolerant to substitution as few amino acid changes were identified and thus are likely to contribute little to 5mC excision activity. Notably, two variable regions (797–1189 and 1406–1484) located between the three conserved domains contained very few critical substitutions (Fig. 3). These two interdomain regions are hereafter named IDR1 and IDR2, respectively.
Fig. 3.
Distribution of random amino acid substitutions that affect 5mC glycosylase activity of DME. The box represents the coding region of full-length DME. An arrow indicates the point of ΔN677 truncation. Vertical bars within the box represent the positions where critical single amino acid changes are observed. Positions of the three conserved domains—domain A (orange), glycosylase domain (magenta), and domain B (green)—are indicated above the box. Tick marks indicate positions every 100 amino acids.
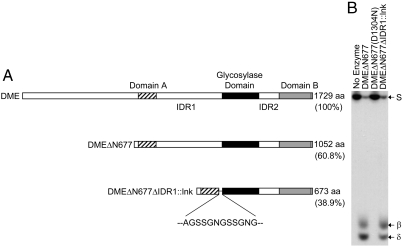
The paucity of amino acids substitutions affecting glycosylase activity in the nonconserved variable regions suggested that we should be able to remove these regions without compromising the enzyme activity. To test this idea, we generated a recombinant DME fragment in which both N-terminal 677 amino acids and IDR1 were removed. Instead, both domain A and the glycosylase domain were tethered together by a short linker peptide (lnk), replacing 393 amino acids of IDR1. As a consequence, we produced a recombinant protein DMEΔN677ΔIDR1∷lnk that consisted of a total of 673 amino acids, whose size was reduced to 38.9% (673/1,729) of full-length DME (Fig. 4A) while maintaining intact three conserved domains. As shown in Fig. 4B, DMEΔN677ΔIDR1∷lnk retained 5mC glycosylase activity, producing both the same β- and δ-elimination products as observed in a reaction with DMEΔN677, which had an intact IDR1. Thus, the large nonconserved IDR1 region is not required for DME activity. This result suggests that both enzymes (DMEΔN677ΔIDR1∷lnk and DMEΔN677) employ the same reaction mechanisms, regardless of the presence or absence of IDR1, to remove 5mC from DNA. This also demonstrates that the three distinct domains—domain A, glycosylase domain, and domain B—are necessary and sufficient for the glycosylase function of DME. Considering that each domain is highly conserved in all DME homologs from various species (12, 30), DMEΔN677ΔIDR1∷lnk is likely to possess all “minimal” essential components that are common to the DME class of 5mC glycosylases. The necessity of the three conserved domains revealed that DME has a unique structure relative to more extensively characterized single domain glycosylases for excision of target bases.
Fig. 4.
Essential modules for 5mC glycosylase activity of DME. (A) From DMEΔN677, the IDR1 is removed and replaced with a short linker sequence (lnk), producing DMEΔN677ΔIDR1∷lnk. Structures of full-length DME, DMEΔN677, and DMEΔN677ΔIDR1∷lnk are aligned together to compare their relative sizes and positions of the three conserved domains. Sizes of the fragments are indicated to the right. (B) DMEΔN677ΔIDR1∷lnk excises 5mC from methylated DNA substrate similarly to DMEΔN677. No-enzyme control or catalytic mutant DMEΔN677(D1304N) does not display 5mC excision activity. Oligonucleotide substrate (S) and β- and δ-elimination products are shown to the right of the panel.
A Mixed Charge Cluster Is Required for DNA Binding and 5mC Excision.
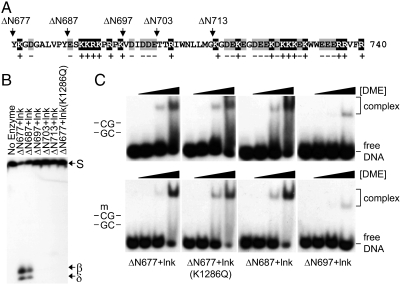
In a random substitution analysis, a total of 18 substitutions were identified in domain A, and all of them were clustered in the second half of the region (amino acids 755–795) (Fig. S3). One interesting feature in the first half is an exceptionally high frequency of both positively and negatively charged residues (18 positive and 17 negative out of 63 residues in region 678–740) (Fig. 5A). Short stretches of basic and acidic residues are present in regions 687–696 and 697–702, respectively, and a long stretch of mixed charge residues follows in region 713–740. Because no deleterious amino acid substitutions were obtained in the first half of the region, we assumed that the overall charge pattern in this area, named as a mixed charge cluster (MCC), is more critical for DME function.
Fig. 5.
Electrophoresis mobility shift assay for truncated DME proteins. (A) Amino acid compositions in domain A (region 678–740) of DME. Positively charged residues (K and R) and negatively charged residues (D and E) are shaded in gray and black, respectively. Positions of deletion are marked by arrows above the sequence of DME. (B) 5mC glycosylase activity of truncated DME proteins. The IDR1 was replaced with a short linker peptide (lnk) in all DME proteins except for ΔN677. The affinity tags were removed from the recombinant proteins and only DME peptides were purified and reacted with methylated oligonucleotide substrate. (C) Binding activity of truncated DME proteins. Increasing amounts of purified DME proteins were incubated with unmethylated (Upper) or methylated (Lower) DNA and separated on a nondenaturing polyacrylamide gel. Relative amount of DME proteins, DME-DNA complex, and free DNA are indicated at the right of the panel.
We made a series of truncations at the N terminus of domain A and generated DMEΔN687-, ΔN697-, ΔN703-, and ΔN713ΔIDR1∷lnk (Fig. 5A). These truncated proteins were purified and tested for 5-mC glycosylase activity. DMEΔN687ΔIDR1∷lnk still possessed glycosylase activity producing signature products in the course of 5mC excision, whereas there was a significant loss of activity with further truncated proteins (Fig. 5B). This result suggests that the MCC in the first part of domain A is necessary for base excision activity of DME. Remarkably, DMEΔN697ΔIDR1∷lnk is devoid of a cluster of positively charged residues (six lysine and arginine residues in the area), which would make the wild-type region highly basic.
In a recent study, a short N-terminal lysine-rich domain of ROS1, a member of the DME family, was reported to mediate DNA binding (30). DME also possesses the same kind of lysine and arginine (KR)-rich domain in the N-terminal region, which was previously identified as a nuclear localization signal (12). Thus, we set out to determine whether a positively charged cluster in domain A is required for DNA binding, which might have a separate function derived from a KR-rich domain in the N-terminal region.
We found that DMEΔN677ΔIDR1∷lnk was able to bind both methylated and unmethylated DNA (Fig. 5C). DNA binding was not affected by a mutation of a catalytic residue as observed in DMEΔN677ΔIDR1∷lnk(K1286Q). These findings suggest that the absence of 5mC has little effect on binding affinity and that DNA binding is independent of the catalytic activity of DME. The same degree of DNA binding persisted in a further truncated DMEΔN687ΔIDR1∷lnk. However, DMEΔN697ΔIDR1∷lnk, in which a positively charged cluster was removed, displayed a significant loss of DNA binding activity (Fig. 5C). Further truncations did not recover DNA binding activity of DME (Fig. S5). These findings suggest that a positively charged cluster in region 688–697 mediates DNA binding in a methylation-independent manner, which should be a prerequisite to 5mC excision in an active site pocket.
Discussion
The DME family proteins have three distinct domains interspersed with poorly conserved regions (Fig. S1), and our results have provided insight into their role in DME activity. A significant portion of the DME N terminus was found to be dispensable for in vitro activity, whereas the C-terminal region was necessary (Fig. S1). Thus, intact regions of all three domains are required for 5mC excision activity of DME. We also confirmed the functional importance of the [4Fe-4S] cluster by site-directed mutagenesis, in which substitutions of some conserved amino acids were found to affect the biochemical activity of DME (Fig. 1). This implies that DME has a similar core structure to other glycosylases such as Endo III and MutY (25) and may utilize a similar base excision mechanism to remove 5mC (31, 32).
Using a random mutagenesis approach, we identified single amino acid substitutions that severely reduced DME activity (Fig. 2 and Table S1). These substitutions provide more detailed structure–function information on the glycosylase domain and the [4Fe-4S] cluster of DME by comparison with other known structures in the HhH superfamily. For instance, two large hydrophobic residues L1277 and V1291, which correspond to L111 and V125 in Endo III, respectively (22, 25), are likely to serve as interhelical packing residues between the two helices of the HhH motif, and their substitutions eliminated the catalytic activity of DME (L1277P and V1291E). In addition, a highly conserved proline residue in the FCL of the [4Fe-4S] cluster, which likely acts as a spacer between positively charged residues for interaction with DNA (25), was proven to be essential for DME because its substitution (P1376L) was critical for activity.
This work revealed a unique structure of DME that consists of a typical HhH-GPD glycosylase domain and two other conserved domains with additional function(s). It is noteworthy that the majority of substitutions found were confined to the three conserved domains, which suggests a distinct modular structure of DME (Fig. 3). A subsequent “cut-and-paste” experiment supported this idea by showing that a recombinant small DME polypeptide in which most of the variable regions had been removed still retained an essential biochemical activity required to excise 5mC (Fig. 4). The lack of a requirement of the interdomain variable regions suggests that the three domains form a globular architecture and not a flexible “beads-on-a-string” arrangement, which has been shown to be important for other multidomain proteins involved in genome maintenance (33, 34).
Why do DME family proteins contain two additional domains flanking the central glycosylase domain? Most DNA glycosylases recognize and remove damaged or modified bases from DNA, which usually exist at a very low frequency in the genome. Therefore, it is a formidable challenge for typical glycosylases to accurately find lesions among a vast number of normal bases (31, 35). However, DME faces the opposite situation—cytosine methylation is highly abundant in the genome, and, therefore, it must either remove a large number of targets or the targets for removal must be selectively chosen (14, 17). In addition to the ability of 5mC to form a strong Watson–Crick base pair with guanine, the presence of the hydrophobic methyl group in the DNA major groove helps to stabilize alternate, non-B-form conformations of DNA (36), which may present another hurdle for the enzyme to overcome. Therefore, two additional domains might be required to resolve the issues mentioned above to excise 5mC in concert with the glycosylase domain. Both domains might have been equipped to help DME family proteins search for lesions by distinguishing structural or thermodynamic differences in C-G and 5mC-G base pairs. In addition, they might function to unwind a double-stranded DNA helix or stabilize a DNA conformation to facilitate base flipping, a mechanism that most DNA glycosylases utilize (20, 37). Further structural and biochemical studies are needed to explore unidentified but essential functions of the two conserved domains.
It was reported that most transcription and replication factors possess one or more charge clusters (38). DME also contains an intriguing charge cluster MCC in “domain A.” We showed that the MCCs mediate nonspecific DNA binding (Fig. 5C). This binding property might be helpful for the enzyme to efficiently locate precise targets, initiate reactions, or stabilize the protein–DNA complex while reactions proceed. Recent studies showed that ROS1, another member of the DME family, locates and excises 5mC in a distributive manner (19) and that a lysine-rich domain at the N terminus increases DNA binding activity (30). However, removal of the N-terminal region did not abolish DNA binding of ROS1, which indicates that a basic region at the N terminus is not directly involved in base excision but rather plays a role in lesion search (30). Therefore, it is assumed that a MCC located in domain A plays a more direct role for base excision possibly by stabilizing DNA–protein contacts at the initiation step of the reaction.
The DME family is highly conserved in diverse plant species, and interestingly, homologs are also present in ancient plant lineages such as Micromonas and Ostreococcus (39). This suggests their primitive roles in a common ancestor of green algae and land plants, probably to cope with a unique cellular environment generated by endosymbiosis events that occurred over a billion years ago. We demonstrated that the three conserved domains are necessary and sufficient for the glycosylase activity in vitro, whereas other variable regions are not required (Fig. 4). This strongly suggests that DME family evolved from a single ancestral gene and that all the functionally essential components have been largely unchanged since their advent. Later duplication and divergence events would expand the repertoire of this family by assigning distinct functions to each member, while retaining essential biochemical activity.
Materials and Methods
Cloning, Protein Expression and Purification, and Glycosylase Activity Assays.
Details are in SI Materials and Methods.
Random Mutagenesis.
In order to construct a vector for random mutagenesis, the full-length DME cDNA was PCR amplified with primers JH-RN3XbaI (5′-GGAATCTAGATACAAAGGAGATGGTGCAC; Xba I site underlined) and JH-Thr-6xHis-SalI (5′-GTCGACTCAATGATGATGATGATGATGGGATCCACGCGGAACCAGGGTTTTGTTGTTCTTC; Sal I site underlined) to introduce Xba I and Sal I sites at the 5′ and 3′ ends of DMEΔN677, respectively, in addition to a thrombin site and 6xHis sequences at the 3′ end. The PCR product was digested with Xba I and Sal I and inserted into the pMAL-c2x vector (NEB) at the corresponding sites, creating the c2x-DMEΔN677-6xHis. A DMEΔN677 fragment was divided into three regions RN, RM, and RC, and each was subjected to random mutagenesis by error-prone PCR using the GeneMorph II Random Mutagenesis Kit (Stratagene). The PCR product was digested with the restriction enzymes and inserted into the c2x-DMEΔN677-6xHis at the corresponding sites replacing a wild-type sequence. The resulting plasmids were transformed into E. coli DH5α strains and transformants were plated on the LB/Glu/Amp medium in the presence of 60 μM IPTG. Cells were grown at 28 °C for 24 h and surviving colonies were picked up for further analysis. For detailed information, see SI Materials and Methods.
DNA Binding Assays.
Standard electrophoresis mobility shift assays were performed to measure the DNA binding activity of various forms of DME proteins. Radio-labeled unmethylated or methylated oligonucleotide substrate was prepared as described above. One hundred nanomolars of oligonucleotide substrate were incubated with varying amounts (0, 8, 40, and 200 ng) of DMEΔN677-, DME(K1286Q)ΔN677-, DMEΔN687-, or DMEΔN697ΔIDR1∷lnk proteins in 10 μL of binding buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Triton X-100, 0.1 mg/mL BSA, 10% glycerol, 10 mM DTT) for 10 min at 23 °C. The reactions were separated on a native polyacrylamide gel (4% acrylamide, 2.5% glycerol, 0.5X Tris-Borate-EDTA buffer). The gel was exposed to X-ray film at -80 °C.
Supplementary Material
Acknowledgments.
We thank James Berger and Scott Gradia at University of California–Berkeley for help with the analysis of the secondary structure of DME and the design of a linker sequence. This study is supported by Grant 500-20090148 and Grant 500-20100140 from the Korea Research Foundation (to J.H.H.) and Grant R01-GM069415 from the National Institutes of Health (to R.L.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014348107/-/DCSupplemental.
References
- 1.Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:590–590. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 2.Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 6.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Hajkova P, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooi SKT, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Wu SC, Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 13.Gong ZH, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 14.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 19.Ponferrada-Marin MI, Roldan-Arjona T, Ariza RR. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruner SD, Norman DPG, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 21.Guan Y, et al. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 22.Kuo CF, et al. Atomic structure of the DNA repair [4Fe-4S] enzyme Endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 23.Labahn J, et al. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 24.Yamagata Y, et al. Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell. 1996;86:311–319. doi: 10.1016/s0092-8674(00)80102-6. [DOI] [PubMed] [Google Scholar]
- 25.Thayer MM, Ahern H, Xing DX, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repairenzyme Endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chepanoske CL, Lukianova OA, Lombard M, Golinelli-Cohen MP, David SS. A residue in MutY important for catalysis identified by photocross-linking and mass spectrometry. Biochemistry. 2004;43:651–662. doi: 10.1021/bi035537e. [DOI] [PubMed] [Google Scholar]
- 27.Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005;9:145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 29.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci USA. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponferrada-Marin MI, Martinez-Macias MI, Morales-Ruiz T, Roldan-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J Biol Chem. 2010;285:23030–23037. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brosey CA, et al. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J Am Chem Soc. 2009;131:6346–6347. doi: 10.1021/ja9013634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson PD, Chagot B, Chazin WJ, Eichman BF. Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif. J Biol Chem. 2010;285:22940–22947. doi: 10.1074/jbc.M110.131276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vargason JM, Eichman BF, Ho PS. The extended and eccentric E-DNA structure induced by cytosine methylation or bromination. Nat Struct Biol. 2000;7:758–761. doi: 10.1038/78985. [DOI] [PubMed] [Google Scholar]
- 37.Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res. 2000;460:201–210. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 38.Brendel V, Karlin S. Association of charge clusters with functional domains of cellular transcription factors. Proc Natl Acad Sci USA. 1989;86:5698–5702. doi: 10.1073/pnas.86.15.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.