Abstract
A unique multicomponent vaccine against serogroup B meningococci incorporates the novel genome-derived proteins fHbp, NHBA, and NadA that may vary in sequence and level of expression. Measuring the effectiveness of such vaccines, using the accepted correlate of protection against invasive meningococcal disease, could require performing the serum bactericidal assay (SBA) against many diverse strains for each geographic region. This approach is impractical, especially for infants, where serum volumes are very limited. To address this, we developed the meningococcal antigen typing system (MATS) by combining a unique vaccine antigen-specific ELISA, which detects qualitative and quantitative differences in antigens, with PorA genotyping information. The ELISA correlates with killing of strains by SBA and measures both immunologic cross-reactivity and quantity of the antigens NHBA, NadA, and fHbp. We found that strains exceeding a threshold value in the ELISA for any of the three vaccine antigens had ≥80% probability of being killed by immune serum in the SBA. Strains positive for two or more antigens had a 96% probability of being killed. Inclusion of multiple different antigens in the vaccine improves breadth of coverage and prevents loss of coverage if one antigen mutates or is lost. The finding that a simple and high-throughput assay correlates with bactericidal activity is a milestone in meningococcal vaccine development. This assay allows typing of large panels of strains and prediction of coverage of protein-based meningococcal vaccines. Similar assays may be used for protein-based vaccines against other bacteria.
Keywords: serogroup B, typing, meningococcal antigen typing system (MATS), bactericidal
In the global effort to eliminate bacterial meningitis and septicemia, serogroup B Neisseria meningitidis is now among the most challenging pathogens for vaccine development (1, 2). Vaccines based on meningococcal serogroups A, C, W135, and Y capsular polysaccharide conjugates have been licensed in many parts of the world (3–6). For serogroup B, only strain-specific outer membrane protein vaccines were developed (7, 8) because the serogroup B capsular polysaccharide is not immunogenic and is a potential autoantigen (9, 10). Both types of vaccine have been evaluated for immunogenicity by means of complement-mediated killing of bacteria in the serum bactericidal assay (SBA), an assay for functional antibodies that was established as a correlate of protection in the 1960s (11–14).
Because protein antigens may vary in their sequence and level of expression, the use in vaccines of novel proteins identified by whole-genome screening creates a challenge for determining which bacteria will be covered by the vaccine. Typing systems would not exist for these antigens, and without an efficient method to assess strain coverage, very large efficacy trials or many functional antibody tests on large panels of isolates would be required. The latter are impractical because of large specimen requirements and the difficulty of scaling up the assays. Solving this problem for serogroup B meningococci could offer solutions for vaccines against other important pathogens such as nontypeable Haemophilus influenzae, Staphylococcus aureus, and Streptococcus groups A and B and pneumoniae.
The vaccine candidate under study [multicomponent meningococcal serogroup B (4CMenB) vaccine] contains four major immunogenic components (15, 16). Two are recombinant fusion proteins: GNA2132 or Neisserial heparin-binding antigen (NHBA) (17, 18) fused with GNA1030, GNA2091 fused with factor H binding protein (fHbp) (19, 20), Neisserial adhesin A (NadA) (21, 22), and outer membrane vesicles (OMV) derived from meningococcal strain NZ98/254. The individual constituents of the fusion proteins and NadA were identified via whole-genome screening, a unique method of antigen discovery (23). The NZ98/254 OMV component, which contains PorA serosubtype 1.4, was effective in a strain-specific outbreak in New Zealand (7, 8).
We report a rapid, reproducible method to define antigen phenotypes that confer susceptibility to killing by the bactericidal antibodies induced by the 4CMenB vaccine. The antigen phenotype depends on both the extent of immunologic recognition and the level of expression of the antigens on different isolates of serogroup B, two important variables that help to determine whether bacteria are killed in the SBA. We found that the vaccine can be expected to cover most MenB strains with fHbp variant 1, or with PorA 1.4, or with sufficient expression of NadA or NHBA.
The inclusion of multiple independent antigens in the vaccine means that strains with multiple vaccine antigens may be more susceptible to SBA, and the overall breadth of strain coverage extended, even if some antigens mutate.
Results
Meningococcal Antigen Typing System (MATS).
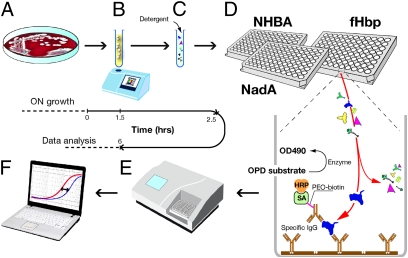
We assessed whether particular bacterial strains express antigens that cross-react with components of the 4CMenB vaccine in a series of assays we have termed the meningococcal antigen typing system (MATS). These assays together identify the phenotype of the individual proteins chosen as vaccine components, fHbp, NadA, NHBA, as well as the genotype of the variable region of PorA, which was used to assess the potential for immunologic recognition of the PorA component. For fHbp, NadA, and NHBA, we developed a sandwich ELISA (Fig. 1) to define phenotypes using antigen capture by purified polyclonal IgG antibodies. We evaluated PorA using genotypic characterization (PCR).
Fig. 1.
Schematic of the MATS ELISA method. (A) MenB bacteria are grown overnight on chocolate agar. (B) A suspension of bacteria taken from the plate is prepared to a specified OD600. (C) Detergent is added to the suspension to extract the capsule and expose the antigens. (D) Serial dilutions of extract are tested in the MATS ELISA. A specific capture antibody (yellow) binds one of the antigens (example: fHbp, blue) from the extract, which is then detected with a specific biotin-labeled antibody (yellow and purple) and a streptavidin–enzyme conjugate (green and gold). (E) Plates are read at 490 nm in an ELISA reader. (F) Results are calculated by comparing the curve of OD490 vs. dilution obtained with the serially diluted unknown strain to a serially diluted reference strain tested in the same ELISA plate.
Relative Potency of NHBA, NadA, and fHbp in Serogroup B Strains.
The MATS ELISA depends on both the quantity of antigen expressed by the bacteria and the extent of its immunologic cross-reactivity with the corresponding antigen in the vaccine. We compared the MATS ELISA reactivity of each N. meningitidis strain to that obtained using a reference MenB strain. The difference in ELISA reactivity, determined mathematically by comparison of serial dilution curves of the two bacterial extracts, was designated the relative potency (RP). In this manner, the RP of fHbp, NadA, and NHBA was measured in 124 serogroup B strains (Table S1) selected to represent a broad range of multi locus sequence type (MLST) and PorA types from varying geographic regions, but not representative of endemic MenB bacteria in any particular country or region. The strains were diverse in their RP values determined by the MATS ELISA, differences that could reflect variations in antigen expression, or in the cross-reactivity of the antigens expressed, or both (Fig. 2). On the basis of MATS, 33 of the 124 strains did not match the vaccine for any of the four major antigens, 41 strains matched the vaccine for a single antigen, 34 for two antigens, and 16 for three antigens.
Fig. 2.
(A–C) Frequency distribution of relative potency (RP) of NHBA, NadA, and fHbp antigens in 124 serogroup B strains. (Upper) Reverse cumulative distributions of the proportion of strains with antigen RP greater than the values indicated on the x axis. (Lower) Histograms of frequency distribution of strains with different values of antigen RP. (A) Antigen RP of different strains for fHbp. Genetic variants of fHbp (variants 1, 2, or 3, with subvariants denoted by decimals) are indicated by brackets. (B) Antigen RP of different strains for NHBA. (C) Antigen RP of NadA in different strains. Brackets denote strains with low expression and higher expression of NadA. Eighty-eight percent of the strains not quantifiable by the test are PCR negative for the NadA gene. rel. pot., relative potency for each antigen determined by MATS in comparison with the reference strains H44/76 (fHbp), 5/99 (NadA), and NGH38 (NHBA). LLOQ, lower limit of quantitation, lowest antigen RP values with between-assay CV ≤20%, 27%, and 34% for fHbp, NHBA, and NadA, respectively.
For fHbp, variations in amino acid sequence had a significant impact on RP (Fig. 2A). Strains with fHbp variant 1.1, which is matched to the vaccine, had the highest RP, the values varying from 46 to 140% of the reference strain. Strains classified within fHbp variant 1, but not subvariant 1.1, showed lower RP, ranging from 1.6 to 38% of the reference strain. In contrast, for strains that expressed fHbp variant 2 or 3, representing 40% of all isolates tested, the RP was below the lower limit of quantitation (LLOQ).
All serogroup B strains possess the gene for NHBA, and 70% of isolates tested had values above the LLOQ, such that RP was from 20% to 130% of that obtained for the reference strain, an approximately sixfold range (Fig. 2B). One third of panel strains possess the gene for NadA; up to 20% gave RP results above the LLOQ (Fig. 2C). RP results for NadA on the panel strains varied over a 1,000-fold range.
Relationship Between MATS and SBA.
To explore the relationship between bactericidal titers and the MATS ELISA for each individual antigen, we performed SBA testing against 57 serogroup B strains from the 124-strain panel using pooled sera from 141 infants who had received three immunizations or three immunizations plus one booster of 4CMenB. To evaluate the performance of MATS in marginal coverage situations, the 57 strains were enriched for those with RP above the LLOQ, but <50% of that of the reference strain for one antigen only, and with RP below the LLOQ for the other three antigens. Fifty-four of the 57 strains were PorA mismatched to 4CMenB. Histograms of MATS ELISA results and a phylogram showing the MLST and MATS types from the 57 strains are shown in Fig. S1.
Among the 57 strains, we selected a subset of 23 strains that, in addition to being mismatched to the vaccine for PorA, had only one vaccine antigen above the LLOQ in the MATS ELISA. In this subset of strains (5 for fHbp, 11 for NHBA, and 7 for NadA) it might be predicted that killing in the SBA would be attributed to only one of the four antigens. In these subsets, we observed higher SBA titers in strains that had higher MATS RP. Despite the limited sample size, the correlations between SBA titers and relative potency were statistically significant (P = 0.005, 0.008, and 0.027 for fHbp, NHBA, and NadA, respectively) (Fig. S2).
To assess the MATS RP as a potential means to estimate strain coverage, we evaluated the panel of 57 strains using a simple criterion of “killed” or “not killed” by 4CMenB on the basis of a fixed SBA titer. Strains were considered killed if pooled sera from infants who received three immunizations plus one booster of 4CMenB achieved an SBA titer ≥8 (if the preimmunization titers were <4) or achieved at least a fourfold rise (if preimmunization titers were ≥4). We found that the point measurement of MATS RP for the majority of strains that were killed in the SBA was greater than a value that we termed the positive bactericidal threshold (PBT). These RP values were 2.1, 29.4, and 0.9% for fHbp, NHBA, and NadA, respectively.
Among strains having a MATS relative potency above the PBT for one or more antigens, 89% were killed in the SBA with pooled serum from 13-mo-old children who were given three immunizations plus one booster of 4CMenB. Among strains with MATS RP at or below the PBT for all antigens, 77% were not killed. The overall accuracy of MATS in predicting whether strains were covered in this age group was 86%, with a statistically significant association (P < 0.0001) between MATS RP and the corresponding SBA result (Table 1). When pooled serum from 7-mo-old infants given three doses of 4CMenB was analyzed, 83% of strains above the PBT were killed and 73% at or below the PBT were not killed, for an accuracy of 80%.
Table 1.
Positive and negative predictive values of MATS for vaccine antigens, based on killing in SBA against a panel of 57 serogroup B strains by pooled sera from 13-mo-old children immunized with 4CMenB at 2, 4, 6, and 12 mo of age
| % positive predictive value* | % negative predictive value† | % accuracy‡ | P value§ | |
| NHBA¶ | 82 (9/11) | 79 | 0.006 | |
| NadA‖ | 83 (5/6) | 79 | 0.024 | |
| fHbp** | 100 (7/7) | 77 (10/13) | 85 | 0.002 |
| Any vaccine antigen or combination of antigens | 89 (39/44) | 86 | <0.0001 |
Shown is the number of strains actually killed in SBA and predicted killed by MATS for the panel of 57 strains and for subpanels in which three of four antigens were negative or at or below the PBT. Each subpanel contained 13 strains with all four antigens at or below the PBT and 7–11 strains with one antigen above the PBT. Individual MATS results for each strain are plotted in Fig. 3.
*Proportion of strains with any of the four antigens above the PBT that are killed in the SBA by immune serum.
†Proportion of strains with all of the four antigens below the PBT that are not killed in the SBA by immune serum.
‡Accuracy, (number of strains predicted killed by MATS and actually killed in SBA + number of strains predicted not killed by MATS and actually not killed in SBA)/total number of strains tested.
§Fisher's exact association test for MATS predicted coverage (any of the four antigens above the PBT) vs. actual SBA killing (titer ≥8) by pooled infant sera.
¶Twenty-four strains with fHbp, NadA, and PorA <PBT.
‖Nineteen strains with fHbp, NHBA, and PorA <PBT.
**Twenty strains with NHBA, NadA, and PorA <PBT.
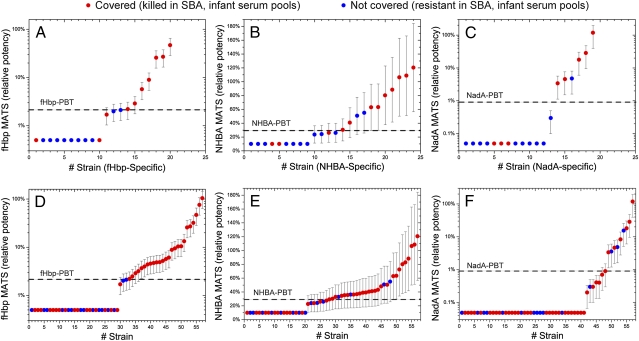
A strain-by-strain comparison of MATS RP with killing of different strains determined in the SBA with serum from immunized human infants is shown in Fig. 3. Fig. 3 A–C shows killing in the SBA of strains that were expected to be targeted by only one of the three major antigens, fHbp, NHBA, or NadA. Fig. 3 D–F shows the complete set of 57 strains ranked by MATS RP, indicating whether they were killed. On the basis of the intermediate precision of the ELISA method (Materials and Methods) we determined 95% confidence limits for the MATS RP measurements, reported as error bars in Fig. 3. In the subset where a single antigen is expected to provide killing, 100% (fHbp), 82% (NHBA), and 83% (NadA) of strains with point measurements of MATS RP greater than the PBT are killed. Nonetheless, overall (Fig. 3 D–F), many strains with a MATS RP at or below the PBT for one antigen are killed, by antibodies to other antigens that are above the PBT.
Fig. 3.
MATS relative potency (RP) of NHBA, NadA, and fHbp determined by the MATS ELISA in antigen-specific subpanels (strains mismatched for the other three antigens, A, B, and C, respectively) and in the 57-strains panel (D, E, and F, respectively). The strains are in ascending order of MATS RP. Strains covered (as determined by killing in the SBA with pooled postimmune infant sera) are shown with solid red circles, and those not covered are shown with solid blue circles. The dashed line represents the positive bactericidal threshold (PBT) for each antigen; this threshold is the relative potency above which at least 80% of MenB strains are killed in SBA with serum from vaccinated subjects. Error bars represent 95% confidence limits for each measurement, deduced from the intermediate precision of the method.
Strain coverage therefore can be defined as the proportion of circulating strains that demonstrate RP above the PBT for at least one vaccine antigen (NadA, NHBA, or fHbp) in the MATS ELISA or are matched to the PorA serosubtype of the OMV component of 4CMenB. Because the intermediate precision of the ELISA method is known, we can determine 95% confidence limits for the RP measurements to estimate vaccine coverage. For the same panel of 57 strains, vaccine coverage predicted by MATS would be 77% [65–88% at 95% confidence limit (CL) determined on the basis of 95% CL for MATS RP] and the actual proportion of strains killed in SBA by the infant serum pools was 74%.
We extended our observations to adolescents immunized with recombinant serogroup B proteins only and to adults immunized with the 4CMenB vaccine. We used a panel of 64 strains, composed of the 57 selected for infants plus 7 additional strains below the PBT for all antigens and excluding PorA-matched strains because the vaccine given to the adolescents did not contain OMV. We found that 67% of these strains were killed by pooled serum from adolescents, and the predicted coverage by MATS was 70% (95% CL, 63–75%). Using the entire panel of 124 strains, 85% were killed in the SBA by pooled serum from adults, and the proportion predicted by MATS to be covered was 72% (95% CL, 55–81%). Fig. 4 shows the relationship between killing in SBA by sera from adults, MATS profile, and clonal complex for the panel of 124 strains.
Fig. 4.
Phylogenetic reconstruction of the MenB strains used in this study. MLST-based phylogenetic reconstruction is shown of the 124 strains that were MATS typed in this study. Color coding shows antigens at or above the PBT. Strains killed in the SBA with postimmune serum from adult vaccinees are coded by a thick black border. Major clonal complexes (CC) are indicated with the founder CC. The phylogenetic tree was obtained with the Neighbor program from the PHYLIP package, with branch lengths computed from the Kimura two-parameter distances.
Advantages of a Multicomponent Vaccine Formulation.
The recombinant serogroup B vaccine with OMV contains multiple antigens, each of which can induce antibodies that can independently kill serogroup B bacteria in the SBA (16). Using serum pools from adults immunized with 4CMenB vaccine in the SBA against the 124 serogroup B strains, we found that among strains with no antigens above the PBT and with a mismatched PorA serosubtype, 22 of 33 (67%) were killed in the SBA (Table 2). In contrast, among the strains with MATS RP above the PBT for one antigen, 35 of 41 (85%) were killed; and for strains with two or more antigens above the PBT, 48 of 50 (96%) were killed. Thus, targeting bacteria with antibodies to multiple antigens increased the proportion of strains killed. As noted above, the negative predictive value of MATS for the recombinant proteins with OMV is <50% in adults, indicating a substantial underestimation of vaccine coverage. Therefore, MATS is a conservative tool for predicting strain coverage in this age group.
Table 2.
Strains killed in the serum bactericidal assay (SBA) by pooled sera from healthy adults who had received three doses of 4CMenB
| Vaccine antigens with relative antigen potency above the positive bactericidal threshold* | No. killed in SBA | Total no. of strains | % (95% CI†) |
| None | 22 | 33 | 67 (48–81) |
| One | |||
| NHBA | 14 | 18 | 78 (52–93) |
| NadA | 5 | 6 | 83 (36–99) |
| fHBP | 16 | 17 | 94 (69–100) |
| Total | 35 | 41 | 85 (70–94) |
| Two | |||
| fHBP and NHBA | 27 | 29 | 93 (76–99) |
| fHBP and PorA | 2 | 2 | 100 (20–100) |
| NHBA and NadA | 3 | 3 | 100 (31–100) |
| Total | 32 | 34 | 94 (79–99) |
| Three | |||
| PorA and fHBP and NHBA | 13 | 13 | 100 (72–100) |
| fHBP and NadA and NHBA | 3 | 3 | 100 (31–100) |
| Total | 16 | 16 | 100 (76–100) |
| Any (one, two, or three) | 83 | 91 | 91 (83–96) |
*Only antigen combinations that apply to one or more strains are included.
†Ninety-five percent CIs were calculated using the prop.test() function, with Yates’ continuity correction, as implemented in the R package, version 2.4.0 (www.r-project.org).
Discussion
The 4CMenB vaccine against serogroup B meningococcus is the prototype of several vaccines, discovered by genome mining, that are now in development. In each case, the classical bacterial typing systems are not useful because the vaccines are based on novel protein antigens that were previously unknown. Therefore, new typing systems need to be developed. Here we describe a unique typing system for the 4CMenB vaccine against meningococcus. This vaccine contains surface proteins that occur across most strains and elicits bactericidal antibodies that can confer protective immune responses (8, 23–26).
In contrast to polysaccharides that are homogeneous and highly conserved, surface proteins are heterogeneous and their level of expression may vary independently of the markers used in other typing methods, complicating the development of protein-based meningococcal vaccines (16). Therefore, we needed to identify a phenotype of the bacteria predictive of the potential for antibodies raised against the unique components of the vaccine, to kill those strains in SBA.
We established a sandwich ELISA that could be performed on a bacterial cell extract as a means of simultaneously assessing both the cross-reactivity and the abundance of the antigens of interest and quantitated this signal by comparing the relative potency of unknown strains with a reference MenB strain selected for high and consistent expression of one vaccine antigen. We found that MATS relative potency could predict whether a strain would be killed in SBA by antibodies to the 4CMenB vaccine. We studied sera from adults in SBA against a panel of 124 strains, sera from adolescents against 64 strains, and sera from 7 and 13 mo olds (from which only very small volumes were available) against a subset of 57 strains selected from the larger panel. Sera from adults and adolescents were selected to be SBA positive against the reference strains H44/76, 5/99, and NZ98/254 and infant sera were randomly selected from vaccine recipients. We found that MATS relative potency reflected the ability of strains to be killed in SBA by pooled immune sera. Taken together with conventional PCR typing of the PorA antigen, we found that the typing system MATS described the ability of different MenB strains to be covered by 4CMenB in the three age groups.
We found the best agreement between killing in the SBA and relative potency in the MATS ELISA if the same polyclonal antibodies were used for capture and detection (Table S2). Therefore, the ability of diverse epitopes on a target antigen to be bound by different antibodies might be important in determining whether a particular sequence variant can be a target for bactericidal antibodies against a given parent antigen. Thus, the antibodies induced by the vaccine might be bactericidal for a sequence variant if it retained the ability to be bound simultaneously in at least two sites by different antibody molecules. Granoff et al. (20, 27) observed that monoclonal antibodies that bound to different sites on fHbp could be bactericidal when combined, even if individually they were not.
Previously, we studied NHBA, fHbp, and NadA individually to establish that each contributed to killing in the SBA (16). In the present study, we used subsets of strains expressing different variants of the vaccine antigens at different levels to establish a threshold value for each antigen for killing of the bacteria (the PBT) and determined the distribution of each antigen within a population of different serogroup B strains. Together, the vaccine antigens provided overlapping coverage of the majority of the serogroup B strains we studied. In the collection of 124 strains that we studied in this paper, 91 strains had at least one antigen at sufficient levels that it could independently account for killing of the strain, and of those, 50 strains had two or more antigens above this level. All but 2 of these 50 strains were killed in the SBA. Targeting bacteria with multiple antigens thus increased the proportion of strains killed and also could provide protection even if any single antigen mutates or is lost on a given strain.
The level of expression of NadA is regulated by the repressor protein NadR and can be growth phase dependent (28). In our studies, care was taken to maintain the bacteria under standardized growth conditions. Nonetheless, we found 11 strains with the NadA gene that had no NadA expression detectable by MATS and 18 further strains that had very low levels of NadA. As growth conditions in vivo likely differ from those in vitro, our results may not necessarily predict the ability of bactericidal antibodies to kill NadA-bearing serogroup B strains during invasive disease. Litt et al. (29) demonstrated that children recovering from invasive meningococcal disease produced specific antibody responses against NadA, indicating that it can be expressed in an immunogenic form during meningoccal infection.
In pooled adult immune sera, some strains were killed in SBA that were not expected to be killed using our MATS methods. This finding could reflect additional antigens in OMV, such as PorB, Opc, FetA, and LOS, for which we did not perform typing. Wedege et al. (8) demonstrated that adults immunized with OMV could mount cross-reactive SBA responses against PorA mismatched strains. Another explanation for this finding is a synergistic effect between vaccine antigens. Giuliani et al. (16) observed that antibodies that were not independently bactericidal could augment the killing effect of bactericidal antibodies against NHBA in the SBA. MATS does not directly consider complementary effects of nonbactericidal antibodies or the effects of minor constituents of OMV and therefore provides a conservative estimate of killing in the SBA. This result was seen with 4CMenB in infants, children, and adults, with 27%, 23%, and 67%, respectively, of strains that were predicted by MATS not to be killed actually being killed in SBA. Fig. 4 and Fig. S1 show the coverage of the 4CMenB vaccine in adults and infants as predicted by MATS and show that the protection is broad and independent of the genotype determined by MLST.
Estimating the potential protective ability of recombinant multicomponent serogroup B vaccines is challenging because of the difficulty of empirical SBA testing in adequate subjects against sufficient strains to achieve adequate statistical power. We found that relative potency determined by the MATS ELISA can be used as a conservative predictor of SBA outcomes for serogroup B vaccines. The SBA itself, although a generally accepted surrogate marker of resistance to meningococcal disease, measures only the total of functional antibodies in a sample and on its own does not provide information about antibody responses to specific antigens or subcomponents of the vaccine (16). For this reason the SBA cannot precisely define how much antibody is produced to each specific component, whether antibodies to different antigens might have synergistic effects, or whether specific vaccine compositions could be tailored to specific MenB populations or age groups. Nevertheless, the MATS approach did succeed in linking killing in the SBA to the presence of specific target antigens on the bacteria, even when the antigens underwent sequence variation and regulated expression.
MATS provides a rapid estimate of strain coverage on the basis of the vaccine components and the relative antigenic potency (immunoreactivity and quantity) of serogroup B strains in a population or region. It also may be possible to use this method to monitor the distribution of vaccine antigens among invasive and carriage isolates of MenB after introduction of the vaccine and to detect the possible emergence of variants resistant to vaccine-induced immune responses. To enable further study of this method we have standardized and transferred it to reference laboratories in the United States and the European Union. This will enable us to evaluate its performance against a much larger collection (>1,500) of national and regional MenB isolates. Methods to measure relative antigenic potency likewise may be a useful tool for evaluating other protein-based vaccines against meningococci and against other species of bacteria that exhibit polymorphisms in both sequence and level of expression of their surface proteins.
Materials and Methods
Bacterial Strains.
One hundred twenty-four serogroup B strains were obtained from meningococcal reference laboratories in the United Kingdom, France, Germany, Italy, Norway, New Zealand, Australia, and the United States. The panel was not intended to be a representative epidemiologic sample of serogroup B either globally or in any particular geographic region. These strains represented a broad range of amino acid sequence variants in NHBA, NadA, and fHbp. Strains were obtained as frozen stocks and were recovered directly onto chocolate agar plates (bioMerieux).
Antibodies.
Rabbit antisera against fHbp, NadA, and NHBA were obtained by immunization of rabbits with purified recombinant proteins produced in Escherichia coli and formulated in aluminum hydroxide adjuvant. Although the vaccine contains a fusion protein for fHbp, the individual protein was used to obtain specific antisera. The same fusion protein of NHBA with genome-derived neisserial antigen (GNA) 1030 that is included in the vaccine was used to immunize rabbits because of its superior immunogenicity. Recombinant NHBA covalently coupled to a solid phase was then used to affinity purify the antibodies to NHBA. NadA was given as a single protein in the same form as used in the vaccine.
Postimmunization Sera Obtained from Human Subjects.
Healthy human adult volunteers were immunized under informed consent with the 4CMenB vaccine containing 50 μg each of GNA 2091-fHbp, NHBA-GNA 1030, and NadA and 25 μg of outer membrane vesicles from the New Zealand strain NZ394/98, adsorbed to aluminum hydroxide. Adolescents were immunized with 50 μg each of the recombinant proteins and aluminum adjuvant only without the OMV component. Serum samples obtained before immunization and after the second (adults) and third (adolescents) dose of vaccine were screened for bactericidal activity against the strains H44/76, 5/99, and NZ394/98. Pools of 10–12 subjects were prepared that were negative in the SBA against these three strains before immunization and positive after the second dose. Infants were immunized with parental informed consent at 2, 4, 6, and 12–13 mo of age and 0.1-mL aliquots of sera from randomly selected groups of 50–100 subjects with unknown SBA status were pooled for testing in the SBA.
Serum Bactericidal Assay.
SBAs were performed using exogenous human complement as described by Borrow et al. (30) Human complement was obtained from volunteer donors under informed consent. Complement donations were screened before use to ensure they lacked endogenous bactericidal activity at concentrations of both 25 and 50% in the SBA and that they supported bactericidal activity. Both serum and plasma complement were used. The activity of plasma complement was reconstituted by the addition of divalent cations immediately before use. Although the original papers by Goldschneider et al. (13) defined a protective titer cutoff of ≥4, we set the threshold for positive killing in the SBA with pooled serum at the more conservative level of ≥8 because each pool was tested in a limited number of replicates.
Antigen-Specific Sandwich ELISA (MATS ELISA).
A diagram depicting the MATS ELISA method is shown in Fig. 1. Stock cultures of serogroup B bacteria were inoculated onto chocolate agar plates (bioMerieux) and incubated at 37 °C with 100% relative humidity and 5% CO2. After 16–18 h, 10–20 selected colonies were resuspended in 4 mL of Mueller–Hinton broth to an OD600 of 0.4. Care was taken to avoid clumping of the bacteria and to standardize the instruments and cuvettetes used to determine the OD. We tested the ability of various detergents to preserve the antigenicity of the proteins while giving most efficient extraction of the capsule and selected the zwitterionic detergent Empigen BB. Empigen BB (5% in 10× PBS with 0.25% proclin as preservative and 0.01% methylene blue) was added to the bacterial suspension to a final dilution of 1:11 (0.45%) and samples were mixed thoroughly to separate capsular polysaccharide. Twofold serial dilutions of bacterial extract in Mueller–Hinton broth with 0.45% Empigen were plated in duplicate in ELISA plates (COSTAR) that were precoated with rabbit polyclonal antibodies against fHbp, NHBA, or NadA. Plates were sealed and incubated for 1 h at 37 °C and then washed with PBS + 0.05% Tween. Plates were then incubated with biotinylated rabbit polyclonal antibody for 1 h at 37 °C, washed, and incubated with streptavidin-HRP (Jackson ImmunoResearch) for 30 min at 37 °C. Plates were developed with OPD (Sigma) for 20 min at room temperature and then reactions were stopped by addition of 50 μL of 4 N H2SO4. Plates were read immediately at 492 nm with a Molecular Devices plate reader. Recombinant protein at a fixed concentration and serial dilutions of a specific reference strain of bacteria for each antigen of interest (H44/76, NGH38, or 5/99) were included on each plate. The relative potency of each unknown strain was calculated with the appropriate reference strain by comparing five-parameter logistic regression curves fit to twofold serial dilutions of extracts from the reference and unknown strains. The reference strains were as follows: for fHbp, H44/76 (fHbp 1.1, ST-32, ET-5, B:15:P1.7,16, NHBA 35.3%, NadA < LLQ, Norway, 1976),;for NadA, 5/99 (fHbp 2.8, ST-1349, ET-34, B:2b:P1.5,2, fHbp and NHBA < LLQ, Norway, 1999); and for NHBA, NGH-38 (fHbp 2.9, ST-36, B:NT:P1.3, fHbp and NadA < LLQ, Norway, 1988). The curves were fit and the relative potency was calculated using the variance-weighted regression method implemented in the StatLIA software (Brendan Technologies). The reference strain for each antigen was assigned an arbitrary value of 100.
Characterization of Test Performance.
We selected 18 strains covering a wide range of responses for each of the three antigens and tested them repeatedly (six or more assays) on different days and by different operators, to determine acceptance criteria, LLOQ, specificity, relative accuracy, and intermediate precision (% CV) of the test. Results are shown in Table S3. From the intermediate precision, we derived 95% confidence intervals for the MATS relative potency as ±1.96 × (% CV). We also used these data to determine the correct weights for the nonlinear weighted regression. The relationship between relative potency and variance was fitted with a power regression for both unknowns and standard samples, and the regression for the unknowns was used to determine weights.
Determination of PBT.
SBA activity can result from antibodies to many surface proteins. To identify a MATS cutoff value for each antigen that best predicts SBA activity, we evaluated all combinations of MATS values for the 57-strain panel (n = 28, 35, and 15 for fHbp, NHBA, NadA, respectively, giving 14,700 combinations), using a stepwise optimization algorithm: (i) meeting overall and antigen-specific positive predictive value (PPV) ≥ 80% was fulfilled by 2,349 of the 14,700 possible combinations of fHbp/NHBA/NadA PBTs; (ii) maximizing overall and antigen-specific negative predictive value (NPV) was fulfilled by 3 of the 2,349 combinations; (iii) maximizing overall and antigen-specific accuracy was fulfilled by 2 of the 3 previously selected combinations; and (iv) selecting the most conservative combination (higher PBTs) gives the final PBT values: 2.1%, 29.4%, and 0.9% for fHbp, NHBA, and NadA, respectively.
Supplementary Material
Acknowledgments
We thank Lisa De Tora for outstanding editorial advice and assistance, Annett Kleinschmidt and Philipp Oster for help in obtaining clinical trial specimens, Dan Granoff and Joanne Welsch (Childrens’ Hospital Oakland Research Institute) for JAR monoclonal antibodies, Simona Toti for statistics support, Emilio Siena for the phylogenetic analyses, Giorgio Corsi for expert preparation of the artwork, and James Wassil for helpful discussions.
Footnotes
J.D., D.M., G.B., A.B., M.S., M.C., S.B., A.M., W.A., J.C., G.S., L.S., P.B., D.S., M.P., R.R., and M.M.G. are employees of Novartis Vaccines and Diagnostics. J.W., C.F., and E.R.M. are paid consultants to Novartis Vaccines.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013758107/-/DCSupplemental.
References
- 1.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 2.Borrow R, et al. Neisseria meningitidis group B correlates of protection and assay standardization—international meeting report Emory University, Atlanta, Georgia, United States, 16–17 March 2005. Vaccine. 2006;24:5093–5107. doi: 10.1016/j.vaccine.2006.03.091. [DOI] [PubMed] [Google Scholar]
- 3.Tan LK, Carlone GM, Borrow R. Advances in the development of vaccines against Neisseria meningitidis. N Engl J Med. 2010;362:1511–1520. doi: 10.1056/NEJMra0906357. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay ME, Andrews N, Kaczmarski EB, Miller E. Efficacy of meningococcal serogroup C conjugate vaccine in teenagers and toddlers in England. Lancet. 2001;357:195–196. doi: 10.1016/S0140-6736(00)03594-7. [DOI] [PubMed] [Google Scholar]
- 5.Costantino P, et al. Development and phase 1 clinical testing of a conjugate vaccine against meningococcus A and C. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410x(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 6.Snape MD, Pollard AJ. Meningococcal polysaccharide-protein conjugate vaccines. Lancet Infect Dis. 2005;5:21–30. doi: 10.1016/S1473-3099(04)01251-4. [DOI] [PubMed] [Google Scholar]
- 7.Oster P, et al. MeNZB: A safe and highly immunogenic tailor-made vaccine against the New Zealand Neisseria meningitidis serogroup B disease epidemic strain. Vaccine. 2005;23:2191–2196. doi: 10.1016/j.vaccine.2005.01.063. [DOI] [PubMed] [Google Scholar]
- 8.Wedege E, et al. Functional and specific antibody responses in adult volunteers in New Zealand who were given one of two different meningococcal serogroup B outer membrane vesicle vaccines. Clin Vaccine Immunol. 2007;14:830–838. doi: 10.1128/CVI.00039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finne J, Bitter-Suermann D, Goridis C, Finne U. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J Immunol. 1987;138:4402–4407. [PubMed] [Google Scholar]
- 10.Finne J, Leinonen M, Mäkelä PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 11.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. II. Development of natural immunity. J Exp Med. 1969;129:1327–1348. doi: 10.1084/jem.129.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotschlich EC, Goldschneider I, Artenstein MS. Human immunity to the meningococcus. IV. Immunogenicity of group A and group C meningococcal polysaccharides in human volunteers. J Exp Med. 1969;129:1367–1384. doi: 10.1084/jem.129.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotschlich EC, Goldschneider I, Artenstein MS. Human immunity to the meningococcus. V. The effect of immunization with meningococcal group C polysaccharide on the carrier state. J Exp Med. 1969;129:1385–1395. doi: 10.1084/jem.129.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuliani MM, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006;103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuliani MM, et al. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine. 2010;28:5023–5030. doi: 10.1016/j.vaccine.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Serruto D, et al. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci USA. 2010;107:3770–3775. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welsch JA, et al. Antibody to genome-derived neisserial antigen 2132, a Neisseria meningitidis candidate vaccine, confers protection against bacteremia in the absence of complement-mediated bactericidal activity. J Infect Dis. 2003;188:1730–1740. doi: 10.1086/379375. [DOI] [PubMed] [Google Scholar]
- 19.Masignani V, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197:789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beernink PT, Granoff DM. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect Immun. 2008;76:2568–2575. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comanducci M, et al. NadA, a novel vaccine candidate of Neisseria meningitidis. J Exp Med. 2002;195:1445–1454. doi: 10.1084/jem.20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capecchi B, et al. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol Microbiol. 2005;55:687–698. doi: 10.1111/j.1365-2958.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 23.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 24.Holst J, et al. Serum bactericidal activity correlates with the vaccine efficacy of outer membrane vesicle vaccines against Neisseria meningitidis serogroup B disease. Vaccine. 2003;21:734–737. doi: 10.1016/s0264-410x(02)00591-1. [DOI] [PubMed] [Google Scholar]
- 25.Tappero JW, et al. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: A randomized controlled trial in Chile. JAMA. 1999;281:1520–1527. doi: 10.1001/jama.281.16.1520. [DOI] [PubMed] [Google Scholar]
- 26.Holst J, et al. The concept of “tailor-made”, protein-based, outer membrane vesicle vaccines against meningococcal disease. Vaccine. 2005;23:2202–2205. doi: 10.1016/j.vaccine.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 27.Welsch JA, Granoff D. Immunity to Neisseria meningitidis group B in adults despite lack of serum bactericidal antibody. Clin Vaccine Immunol. 2007;14:1596–1602. doi: 10.1128/CVI.00341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metruccio MM, et al. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog. 2009;5:e1000710. doi: 10.1371/journal.ppat.1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litt DJ, et al. Putative vaccine antigens from Neisseria meningitidis recognized by serum antibodies of young children convalescing after meningococcal disease. J Infect Dis. 2004;190:1488–1497. doi: 10.1086/424464. [DOI] [PubMed] [Google Scholar]
- 30.Borrow R, et al. Interlaboratory standardization of the measurement of serum bactericidal activity by using human complement against meningococcal serogroup b, strain 44/76-SL, before and after vaccination with the Norwegian MenBvac outer membrane vesicle vaccine. Clin Diagn Lab Immunol. 2005;12:970–976. doi: 10.1128/CDLI.12.8.970-976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.