Abstract
Chloroplasts move toward weak light (accumulation response) and away from strong light (avoidance response). The fast and accurate movement of chloroplasts in response to ambient light conditions is essential for efficient photosynthesis and photodamage prevention in chloroplasts. Here, we report that two Arabidopsis mutants, weak chloroplast movement under blue light 1 (web1) and web2, are defective in both the avoidance and the accumulation responses. Map-based cloning revealed that both genes encode coiled-coil proteins and that WEB2 is identical to the plastid movement impaired 2 (PMI2) gene. The velocities of chloroplast movement in web1 and pmi2 were approximately threefold lower than that in the wild type. Defects in the avoidance response of web1 and pmi2 were suppressed by mutation of the J-domain protein required for chloroplast accumulation response 1 (JAC1) gene, which is essential for the accumulation response; these results indicate that WEB1 and PMI2 play a role in suppressing JAC1 under strong light conditions. A yeast two-hybrid analysis and a nuclear recruitment assay identified a physical interaction between WEB1 and PMI2, and transient expression analysis of CFP-WEB1 and YFP-PMI2 revealed that they colocalized in the cytosol. Bimolecular fluorescence complementation analysis confirmed the interaction of these proteins in the cytosol. Blue light-induced changes in short chloroplast actin filaments (cp-actin filaments) were impaired in both web1 and pmi2. Our findings suggest that a cytosolic WEB1–PMI2 complex maintains the velocity of chloroplast photorelocation movement via cp-actin filament regulation.
Keywords: Arabidopsis thaliana, chloroplast velocity, phototropin, protein–protein interaction
Chloroplasts capture sunlight for energy production through photosynthesis. To optimize performance, the intracellular locations of chloroplasts change in response to different light conditions. Chloroplasts move toward low-intensity light, resulting in accumulation along the periclinal walls to maximize light capture (the accumulation response) (1). In addition, they avoid high-intensity light, resulting in accumulation along the anticlinal walls to reduce photodamage by high-intensity light (the avoidance response) (2). Recent studies have identified several genes involved in blue light-induced chloroplast movement in Arabidopsis (1). The blue light receptors, phototropin 1 (phot1) and phot2, redundantly mediate the accumulation response (3), whereas phot2 alone mediates the avoidance response (4, 5). Phototropins also regulate blue light-induced chloroplast movement in ferns and mosses and probably in green algae (1). Chloroplast Unusual Positioning 1 (CHUP1), an actin- and profilin-binding protein, localizes to the chloroplast outer membrane (6–8) and functions in chloroplast photorelocation movement and attachment to the plasma membrane (2, 6, 7, 9). More recently, we identified two proteins, Kinesin-like Protein for Actin-Based Chloroplast Movement 1 (KAC1) and KAC2, that are essential for chloroplast photorelocation movement and attachment to the plasma membrane (10). Other proteins involved in chloroplast photorelocation movements have also been identified, including J-domain protein required for chloroplast accumulation response 1 (JAC1), which is involved in the accumulation response and dark positioning (11), plastid movement impaired 1 (PMI1) (12), and a long coiled-coil protein, plastid movement impaired 2 (PMI2) (13). However, the functions of these proteins remain to be elucidated.
To achieve an immediate and accurate response to fluctuating ambient light conditions, a precise regulation of chloroplast velocity is required. The velocity of the chloroplast avoidance movement is dependent on the light intensity and the amount of phot2 (i.e., the higher the light intensity, the faster the velocity of chloroplast movement). The chloroplast velocity in the PHOT2/phot2 heterozygous mutant was half that of WT (14). Simultaneous irradiation with red light enhanced the blue light-induced chloroplast accumulation movement, possibly by increasing cytoplasmic motility (15). Recently we identified chloroplast actin (cp-actin) filaments that are involved in chloroplast photorelocation movement and positioning (9). The presence of cp-actin filaments at the interface between the chloroplast and the plasma membrane depends on CHUP1, KAC1, and KAC2 (9, 10). Blue light irradiation induces the relocalization of cp-actin filaments to the leading edge of chloroplasts before and during photorelocation (which is mediated by phototropins) (9). As the light intensity increases, the difference in the abundance of cp-actin filaments between the front and rear halves of chloroplasts also increases, indicating that light regulates the abundance of cp-actin filaments and thereby determines the velocity of chloroplast avoidance movement (9). Abundance of KAC proteins positively correlated with cp-actin filament amount and the velocity of chloroplast avoidance movement (10). The mechanism and other factor(s) that regulate the velocity of chloroplast movement remain to be determined.
In this study, we identified two Arabidopsis mutants, weak chloroplast movement under blue light 1 (web1) and web2/pmi2, in which the chloroplasts moved more slowly than those of WT. Blue light-induced regulations of cp-actin filaments were impaired in both mutants. WEB1 and PMI2 are coiled-coil proteins that interact in the cytosol. Our data suggest that a WEB1–PMI2 complex maintains the velocity of chloroplast movements via the regulation of cp-actin filaments.
Results
Identification of WEB1 and WEB2/PMI2.
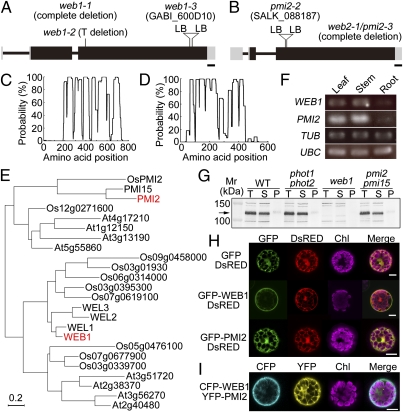
Two Arabidopsis mutants that are defective in chloroplast avoidance response, phot2 and chup1, were previously isolated with a white band assay (WBA) screen (4, 6). During the mutant screen of fast neutron- and γ-ray–mutagenized seeds, we found two complementation groups of the mutant lines that seemed to be deficient in the avoidance response: two web1 mutants [web1-1 (fast neutron) and web1-2 (γ-ray)] and one web2 mutant [web2-1 (γ-ray)]. Because these mutants showed very similar phenotypes (see below), we hypothesized that the same pathway was disrupted in both mutants. Map-based cloning revealed that the WEB1 and WEB2 genes were At2g26570 and At1g66840, respectively (Fig. 1 A and B and Fig. S1 A and B). WEB2 was previously identified as the PMI2 gene (13). Accordingly, web2-1 was renamed pmi2-3. The WEB1 gene has two introns and encodes a protein of 807 amino acids with a predicted molecular mass of ca. 89 kDa (Fig. 1A). The PMI2 gene has two introns and encodes a protein of 607 aa with a predicted molecular mass of ca. 70 kDa (Fig. 1B). The transferred DNA (T-DNA) tagged lines, web1-3 (Fig. 1A) and pmi2-2 (13) (Fig. 1B), showed the same phenotypes as web1 and pmi2, respectively. Expression of WEB1 and PMI2 in the transgenic web1 and pmi2 mutant plants, respectively, rescued the deficient chloroplast movement responses, confirming that At2g26570 and At1g66840 are the genes affected in the web1 and pmi2 mutant plants, respectively. Both WEB1 and PMI2 are predicted to be proteins with high coiled-coil content by the Coils program (16) (Fig. 1 C and D), and they belong to the plant-specific DUF827 (DUF: Domain of Unknown Function; Pfam PF05701) protein family. Arabidopsis and rice have 14 and 10 DUF827 family coiled-coil proteins, respectively (Fig. 1E). Phylogenetic analyses revealed that these proteins could be separated into four clades and that Arabidopsis has three WEB1-like genes (WEL1, -2, and -3) and a PMI2-like gene (PMI15) (13) (Fig. 1E and Fig. S2A). Luesse et al. reported a structure for the PMI2 protein based on in silico analyses and predicted two putative nuclear localization signals (NLS) and a putative ATP/GTP-binding motif A (P-loop) (13) (Fig. S2A). Within the WEB1 sequence, a putative NLS was also predicted, but a P-loop region was not found (Fig. S2A).
Fig. 1.
Identification and subcellular localization of WEB1 and PMI2. (A and B) Schematic illustration of the (A) WEB1 and (B) WEB2/PMI2 genes. The rectangles are exons (gray rectangles indicate the 5′ and 3′ UTRs), and the intervening bars indicate introns. Mutation sites are also indicated. LB, left border of the T-DNA. (Scale bars, 100 bp.) (C and D) Prediction of coiled-coil regions of WEB1 and PMI2. Coiled-coil regions of (C) WEB1 and (D) PMI2 were predicted using the Coils program (http://www.ch.embnet.org/software/COILS_form.html). (E) A rooted phylogenetic tree of WEB1 and PMI2 with related proteins. Arabidopsis (At) and rice (Os) sequences were obtained from the National Center for Biotechnology Information. (F) RT-PCR analysis for gene expression of WEB1 and PMI2. Total RNA was extracted from leaves, stems, and roots of plants and used for RT-PCR. The β-tubulin 2 (TUB) and ubiquitin-conjugating enzyme (UBC) genes were used as controls. (G) Immunoblot analysis of WEB1. Total protein extracts (T) were prepared from the rosette leaves of WT, phot1phot2, web1, and pmi2pmi15 mutants plants and then fractionated into soluble (S) and microsomal pellet (P) fractions by ultracentrifugation (100,000 x g) for 1 h at 4 °C. Immunoblot analysis was performed with an anti-WEB1 polyclonal antibody. The arrow indicates the WEB1 protein. Mr, molecular mass markers. (H) Subcellular localization analysis using GFP fusion proteins. GFP-WEB1 or GFP-PMI2 and the DsRED monomer, as a cytosolic control, were introduced into Arabidopsis protoplast cells via PEG-mediated transformation. Fluorescence images of GFP, DsRED, and chlorophyll autofluorescence (Chl) were captured with a confocal microscope (SP5; Leica) and are shown in green, red, and magenta, respectively. Merged images were constructed using the Leica Application Suite Advanced Fluorescence software. GFP alone was used as a control. (Scale bars, 10 μm.) (I) Colocalization analysis using CFP and YFP fusion proteins. CFP-WEB1 and YFP-PMI2 were coexpressed in Arabidopsis protoplasts. Images of CFP, YFP, and chlorophyll autofluorescence (Chl) were captured and are shown in cyan, yellow, and magenta, respectively. (Scale bars, 10 μm.)
To analyze the WEB1 and PMI2 gene expression patterns, RT-PCR analysis was performed. The β-tubulin 2 (TUB) and ubiquitin-conjugating enzyme (UBC) genes were used as controls. WEB1 and PMI2 were expressed mainly in chloroplast-containing tissues (leaf and stem) and slightly in the roots (Fig. 1F). Gene expression data obtained from the public microarray database Genevestigator (17) revealed that WEB1 and PMI2 are ubiquitously expressed in all tissues (Fig. S2B). Although PMI15 and PMI2 were expressed at similar levels, the three WEL genes were expressed at much lower levels compared with WEB1 in all tissues except for stamen and pollen (Fig. S2B).
Subcellular Localization of WEB1 and PMI2.
Immunoblot analysis with an anti-WEB1 antibody showed that a band of ≈120 kDa was detected in WT plants but not in web1 mutant plants (Fig. 1G and Fig. S3A). This result demonstrated that the molecular mass of the WEB1 protein in vivo was much larger than its predicted molecular mass (ca. 89 kDa). WEB1 proteins expressed in the wheat germ cell-free expression system (Fig. S3B) and in Escherichia coli (Fig. S3C) were also approximately 120 kDa, indicating that the difference in the observed WEB1 molecular mass is not the result of plant-specific posttranslational modification. Although no PMI2 antibody is currently available, hexahistidine-tagged PMI2 proteins expressed in the wheat germ cell-free expression system were about 74 kDa (Fig. S3D), which is similar to the predicted molecular weight (ca. 70 kDa). Immunoblot analysis with fractionated protein samples showed that WEB1 is predominantly detected in the soluble fraction and slightly in the microsomal fractions (Fig. 1G). In the pmi2pmi15 and phot1phot2 double mutants, the amount and fractionation profile of WEB1 were similar to those in WT (Fig. 1G), indicating that the expression amount and localization of the WEB1 protein are not affected by the phototropins or the PMI2 and PMI15 proteins.
Subcellular localization analyses using CFP, GFP, and YFP were performed. The corresponding vectors were transiently coexpressed in Arabidopsis protoplast cells after PEG-mediated transformation (18). The DsRed monomer (DsRED; Clontech) was used as a cytosolic marker. Unexpectedly, GFP-WEB1 was localized predominantly at the cell periphery and slightly in the cytosol (Fig. 1H, Middle), whereas GFP-PMI2 was localized in the cytosol, similarly to DsRED (Fig. 1H, Bottom). Although NLSs were predicted in both the WEB1 and the PMI2 sequences (Fig. S2B), the GFP fusions did not localize in the nucleus, suggesting that the predicted NLSs are not functional (Fig. 1H). To determine whether WEB1 and PMI2 colocalize, CFP-WEB1 and YFP-PMI2 were coexpressed in Arabidopsis protoplasts. Although CFP-WEB1 was mainly localized at the cell periphery, CFP-WEB1 and YFP-PMI2 colocalized in the cytosol (Fig. 1I).
Physical Interaction Between WEB1 and PMI2.
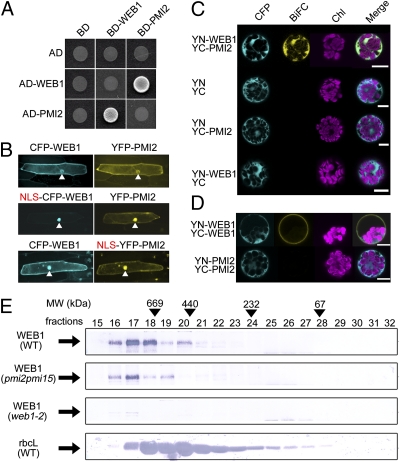
We hypothesized that WEB1 and PMI2 could physically interact for three reasons: (i) the web1 and pmi2 mutants showed the same phenotypes as web1pmi2 double mutant (see below), (ii) coiled-coil regions mediate protein–protein interactions, and (iii) both proteins colocalized in the cytosol when transiently overexpressed. The interaction between WEB1 and PMI2 was verified by yeast two-hybrid (Y2H) analysis. When full-length WEB1 and PMI2 were fused to the Gal4 DNA-binding domain (BD) and the Gal4 activation domain (AD), respectively, and coexpressed in yeast cells, an interaction between BD-WEB1 and AD-PMI2 was clearly observed (Fig. 2A). The result was also confirmed with the BD-PMI2 and AD-WEB1 fusion proteins (Fig. 2A). To clarify the interaction between WEB1 and PMI2 in planta, we took advantage of two different colored fluorescent proteins, CFP and YFP, and an NLS in reference to the previous studies (19, 20), and termed this procedure a nuclear recruitment (NR) assay. The NR assay is based on a recruitment of a cytoplasmic YFP fusion protein into a nucleus by interaction with an NLS-tagged CFP (NLS-CFP) fusion protein that localizes in the nucleus. When the both proteins physically interact, the YFP fusion protein is transported into a nucleus from the cytoplasm by a facilitation of the NLS-CFP fusion protein; thereby both CFP and YFP fluorescence could be detected in the nucleus. As a control experiment, CFP-WEB1 and YFP-PMI2 coexpressed in onion epidermal cells did not localize in the nuclei (Fig. 2B, Top). When NLS-tagged CFP-WEB1 (NLS-CFP-WEB1) and YFP-PMI2 were coexpressed, both proteins were clearly localized in the nucleus (Fig. 2B, Middle). Nuclear localization of CFP-WEB1 was also found in cells that expressed both CFP-WEB1 and NLS-tagged YFP-PMI2 (NLS-YFP-PMI2) (Fig. 2B, Bottom). These results indicate that WEB1 and PMI2 physically interact in planta. To analyze the subcellular localization of the WEB1-PMI2 complex, bimolecular fluorescence complementation (BiFC) analysis (21) with split YFP was carried out. YFP fluorescence was reconstituted by the interaction between YN-WEB1 and YC-PMI2 in the cytosolic fraction of both Arabidopsis mesophyll protoplasts (Fig. 2C) and onion epidermal cells (Fig. S4A). Fluorescence complementation in the cytosol was also observed with the combination of YN-PMI2 and YC-WEB1 (Fig. S4A). Interestingly, YN-WEB1 and YC-WEB1 complemented YFP fluorescence only at the cell periphery but not in the cytosol (Fig. 2D), indicating WEB1 self-association in vivo. On the other hand, YN-PMI2 and YC-PMI2 did not exhibit any fluorescence (Fig. 2D). Because the self-association of WEB1 was not observed by Y2H analysis (Fig. 2A), linker molecule(s) may mediate the WEB1 multimer formation at the cell periphery. Because phot1 and phot2 localize to the plasma membrane (22, 23), one possibility is that phot1 or phot2 is the linker. Another possibility is that PMI2 acts as the linker because PMI2 interacts with WEB1. However, a clear BiFC between YN-WEB1 and YC-WEB1 was observed at the cell periphery in both phot1phot2 and pmi2-3pmi15-1 mutant cells (Fig. S4B), indicating that the phototropins and PMI2 (and PMI15) are not the linker proteins. Overall, the WEB1–PMI2 and WEB1–WEB1 complexes are localized in the cytosol and at the cell periphery, respectively. Consistent with the ability of WEB1 to interact with PMI2 or itself, size exclusion chromatographic analysis of soluble proteins showed that native WEB1 was detected as a large molecular mass complex that is distinct from the rubisco large complex (rbcL) (ca. 550 kDa) (Fig. 2E), indicating that the native WEB1 protein forms large protein complex(es) in vivo. The profile of the WEB1 complex in pmi2pmi15 mutant plants was not significantly different from that in WT, indicating that the WEB1–PMI2 complex is less abundant than the WEB1 homomultimer (Fig. 2E).
Fig. 2.
Physical interaction between WEB1 and PMI2. (A) Y2H analysis. The Gal4 DNA BD and AD fusion proteins were coexpressed in diploid yeast cells that were constructed via mating. The resulting transformants were used in experiments for complementation of growth on SD agar supplemented with a mixture of the appropriate amino acids without Trp, Leu, and His. (B) NR assay with CFP and YFP fusion proteins. Onion epidermal cell layers were bombarded with gold particles coated with the appropriate constructs. After incubation for 24 h in the dark, the cells were observed under a microscope equipped with a fluorescence module (Imager; Carl Zeiss). Arrowheads indicate nuclei. (C) BiFC assay in Arabidopsis protoplast cells. The appropriate BiFC constructs were introduced into Arabidopsis protoplasts via PEG-mediated transformation. After 24 h incubation in the dark, the cells were observed under a confocal microscope (SP5; Leica). (Scale bars, 10 μm.) (D) Detection of WEB1 self-association by BiFC assay. YN-WEB1/YC-WEB1 and YN-PMI2/YC-PMI2 were expressed in Arabidopsis protoplasts. The cells were observed under a confocal microscope (SP5; Leica). (E) Size exclusion chromatographic analysis of the WEB1 protein. Total soluble proteins were subjected to size exclusion chromatography. Individual fractions from a Superdex 200 10/300 column connected to an ÄKTAexplorer 10S were analyzed for WEB1 and rbcL (ca. 550 kDa) by immunoblot analysis with anti-WEB1 and anti-rbcL polyclonal antibodies, respectively. The fraction numbers and the elution peaks of the standard proteins (arrowheads) used for calibration are shown at the top: thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), and BSA (67 kDa).
Both the web1 and the pmi2 Mutants Showed Similarly Slow Chloroplast Movement.
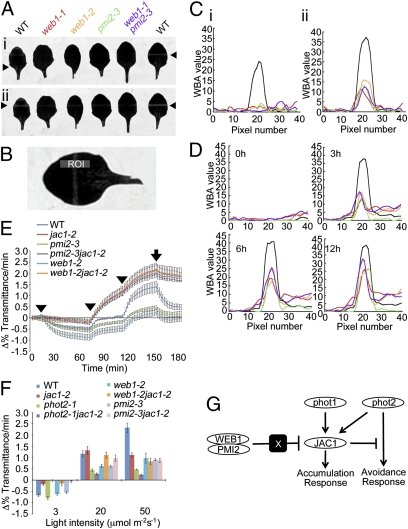
To assess the functions of WEB1 and PMI2 in chloroplast photorelocation movement, the movement was quantitatively examined in web1 and pmi2 single-mutant plants and in a web1pmi2 double-mutant plant (Fig. 3A). Detached rosette leaves from WT, web1-1, web1-2, pmi2-3, and web1-1pmi2-3 plants were examined by WBA. To compare precisely the results obtained from the WBA, a method for WBA evaluation, which is based on quantification of leaf transmittance alteration, was developed. A preliminary test of the method was performed using phot1phot2 mutant leaves (3, 4) (Fig. S5A). The blue channel image was isolated from the RGB color photograph to detect a clear white band on the leaves (Fig. 3A). To quantify the leaf transmittance alterations, a pixel profile of the indicated region of interest (ROI; 10 × 40 pixels) was obtained (Fig. 3B) (see details in SI Materials and Methods). In the resulting graph, the peak of the chart around pixel 20 on the horizontal axis shows the relative intensity of the generated white band, which was termed the WBA value (Fig. 3C and Fig. S5A). A higher WBA value indicates a higher chloroplast avoidance movement response. After irradiation with 500 μmol m−2 s−1 of white light for 30 min, a white band was generated in WT leaves but not in mutant leaves (Fig. 3Ai). The WBA value for WT leaves was ≈25, whereas those of all mutants tested were below the baseline level of 5 (Fig. 3C). Subsequently, irradiation with 1,200 μmol m−2 s−1 of white light for 30 min was tested. White bands were generated in WT and in all mutant plant leaves (Fig. 3Aii), and WBA values of 37.5 and ≈15, respectively, were observed (Fig. 3C). The WBA value for the web1-1pmi2-3 double-mutant plants was the same as those for the single-mutant plants (Fig. 3C), suggesting that WEB1 and PMI2 act in the same pathway. After irradiation with 500 μmol m−2 s−1 of white light for 3, 6, and 12 h, white bands were also observed in web mutant leaves (Fig. 3D and Fig. S5B), and the WBA values increased with irradiation time (Fig. 3D). These results indicate that white bands in web1 and pmi2 mutant plants are not induced to the same extent as in WT, which suggests that the chloroplast movements in both web1 and pmi2 are somewhat attenuated.
Fig. 3.
Phenotypic analysis of web1 and pmi2 mutants and genetic interactions of WEB1 and PMI2 with JAC1. (A) Detached rosette leaves of WT and mutants were aligned on a 0.8% agar plate, covered with a non–light-transmitting black plate with a 1-mm–wide open slit and irradiated with continuous white light (i: 500 μmol m−2 s−1; ii: 1200 μmol m−2 s−1) for 30 min in a growth chamber. Arrowheads indicate the irradiated position. (B) Development of an evaluation method for the WBA. The blue channel image was isolated from an RGB photograph of the leaf after strong light irradiation, and a clear white band on the leaf was detected. The midpoints (pixel number 20) of the long sides of the ROI (10 × 40 pixels) corresponded to the midpoint of the width of the irradiated area, such that the lines on the long sides were perpendicular to the irradiated area. The profile was then measured in the rectangular ROI. (C) Results of the evaluation of the WBA in A. Vertical and horizontal axes show the WBA values and pixel numbers of the long side of the ROI, respectively. (D) Results of the evaluation of the WBA in the time course experiment; detached rosette leaves of WT and mutants were irradiated with a continuous strong white light (500 μmol m−2 s−1) for 0, 3, 6, and 12 h (see also Fig. S5B). (E) Changes in leaf transmittance caused by chloroplast photorelocation movement. Graph shows data from a representative experiment using eight leaves of each indicated genotype. After 10 min in darkness, leaves were irradiated with blue light at 3, 20, and 50 μmol m−2 s−1 sequentially at 10, 70, and 110 min, respectively, as indicated by arrowheads. The blue light was turned off at 150 min (arrow). (F) Speed of the leaf transmittance change over 2–6 min after blue light irradiation. Data were means of four (WT and jac1-2) or three (other mutants) independent experiments. Bars indicate SEs. (G) Possible model of genetic and physical interactions between the proteins involved in chloroplast photorelocation movement. Under strong light condition, both phot1 and phot2 are activated. Although phot2 activate the signaling pathway for chloroplast avoidance response, phot1 (and also phot2) activate the signaling pathway for the accumulation response dependent on JAC1. JAC1 mediates the accumulation response and represses the signaling pathway for chloroplast avoidance response. To promote the avoidance response, WEB1–PMI2 complex prevents JAC1 function via unidentified factor X in cytosol. White ovals and black box indicate the proteins and unidentified factor X, respectively.
Impaired Chloroplast Avoidance Response in web1 and pmi2 Is Dependent on JAC1.
To genetically dissect the roles of the WEB1 and PMI2 genes in chloroplast photorelocation movement, we have analyzed their genetic interactions with the PHOT1, PHOT2, and JAC1 genes, which are involved in chloroplast relocation movements. We found genetic interactions of WEB1 and PMI2 with the JAC1 gene. Chloroplast photorelocation movement was analyzed by leaf transmittance analysis, as previously reported (12, 13), in web1-2jac1-2 and pmi2-3jac1-2 double mutants and compared with those of WT, jac1-2, phot2-1, phot2-1jac1-2, web1-2, and pmi2-3 (Fig. 3 E and F). The jac1 mutant is impaired specifically in the accumulation response but not in the avoidance response (Fig. 3 E and F; note that the speed of the leaf transmittance change in jac1-2 was slower than that of WT under 50 μmol m−2 s−1 blue light), suggesting that JAC1 functions specifically in the accumulation response signaling pathway (11). If WEB1 and PMI2 function in the signaling pathway for the avoidance response, the web1jac1 and pmi2jac1 double mutants would be impaired in both the accumulation and the avoidance responses, like the phot2jac1 double mutant (11). The leaf transmittance changes in response to high fluence rates (20 and 50 μmol m−2 s−1) were smaller in web1-2 and pmi2-3 than in WT (Fig. 3 E and F). Unlike phot2-1jac1-2 (Fig. 3F), both web1-2jac1-2 and pmi2-3jac1-2 showed leaf transmittance kinetics similar to jac1-2 (Fig. 3 E and F), indicating that the jac1 mutation suppressed the web1 and pmi2 mutant phenotypes. In other words, JAC1 repressed the avoidance response in the web1 and pmi2 mutants; accordingly, their chloroplasts exhibited a weak avoidance response (Fig. 3 F and G). Like JAC1 gene, PHOT1 gene is involved only in accumulation response, but not avoidance response (3–5). However, the phot1 mutation did not suppress the web1 and pmi2 mutant phenotypes (Fig. S6A). Thus, both WEB1 and PMI2 precisely maintain chloroplast movement velocity via suppression of JAC1 activity, but not phot1, in WT plants under strong light conditions (Fig. 3G). Although physical interactions of WEB1 and PMI2 with JAC1 were also checked by Y2H assay, the interactions were not observed (Fig. S6B), suggesting that WEB1 and PMI2 suppress the JAC1 activity via unidentified factor (Fig. 3G). A possible model to overview the genetic and physical interactions between the proteins involved in chloroplast photorelocation movement is shown in Fig. 3G.
Impairment of both the Chloroplast Avoidance and the Accumulation Responses.
To directly analyze chloroplast photorelocation movements in these mutants under a microscope, partial cell irradiation with a blue light microbeam was performed on mesophyll cells (15). Chloroplasts in the web1 and pmi2 mutant cells moved away from the strong blue light (Movies S1, S2, S3, S4, and S5), but their velocities were approximately threefold slower than that of WT (Fig. S7A). The velocity of chloroplast movement in the web1wel1wel2wel3 quadruple mutant (Fig. S7B) did not differ significantly from that of the web1-2 single mutant (Fig. S7C), indicating that the WEB1 gene plays the predominant role in chloroplast movement compared with the other WEL genes. Unexpectedly, the accumulation response was also defective in web1 and pmi2 mutants, and their velocities were threefold slower than that of WT (Fig. S7D). These results indicate that both the web1 and the pmi2 mutants have much slower chloroplast velocities in both the avoidance and the accumulation responses.
phot2 Amount and Cytoplasmic Motility.
The amount of phot2 and red light-induced cytoplasmic motility was carefully studied in these mutant plants because these factors were previously reported to affect the velocity of chloroplast movement (14, 15). Immunoblot analysis showed that the amount and membrane localization of the phototropins did not change in web1 and pmi2pmi15 mutants (Fig. S8A). Red light-induced cytoplasmic motility was normal in web1pmi2 mutants (Fig. S8B). Notably, previous studies showed that pmi2 retained normal mitochondrial movement (13), and pmi2pmi15 double mutants showed normal accumulation and fractionation profile of KAC proteins regulating the velocity of chloroplast movement via cp-actin filaments (10). Thus, WEB1–PMI2 complex maintains the chloroplast movement velocity by a mechanism other than those described above.
Defective cp-actin Filament Dynamics During the Chloroplast Avoidance Response in web1 and pmi2 Mutant Cells.
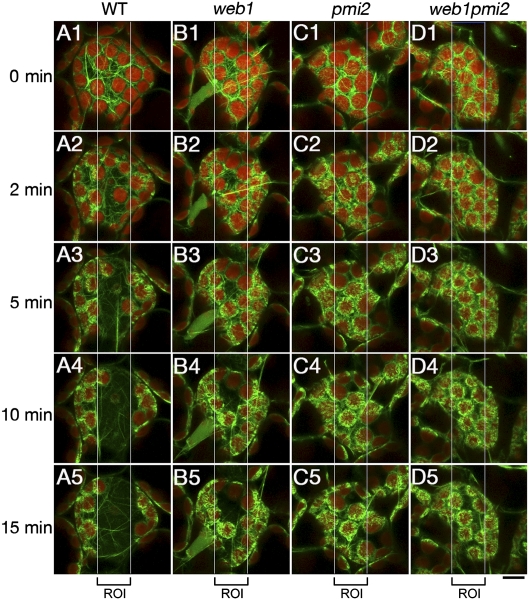
When irradiated with strong blue light to induce the avoidance response in chloroplasts, cp-actin filaments relocalize to the leading edge of moving chloroplasts, and this process is regulated by the phototropins (9). Because the difference in the cp-actin filament amounts between the front and rear ends of moving chloroplasts determines the velocity of chloroplast movement, we analyzed the dynamics of cp-actin filaments during chloroplast avoidance movement. In WT plants, whole cp-actin filaments inside the ROI disappeared after irradiation with strong blue light and then reappeared only at the leading edge of moving chloroplasts outside the ROI (Fig. 4 A2–A5). In contrast, cp-actin filaments were constitutively present in the web1, pmi2, and web1pmi2 mutant cells and did not disappear, even inside the ROI, after irradiation with strong blue light (Fig. 4 B2–D5). These results suggest that cp-actin dynamics are defective in web1 and pmi2 mutant cells.
Fig. 4.
Blue light-induced cp-actin dynamics. (A–D) cp-actin filaments were visualized in the palisade mesophyll cells of WT (A1–A5), web1 (B1–B5), pmi2 (C1–C5), and web1pmi2 (D1–D5) plants. The mesophyll cells expressing GFP-tagged talin (the F-actin binding domain of mouse talin) were observed using a confocal microscope (SP5; Leica). Time-lapse images of cp-actin filaments were captured at the indicated times. Chloroplast avoidance responses were induced by scanning the rectangular ROIs (15 μm × 50 μm) with 2.8 μW light from a 458-nm laser between intervals. The enlarged images covered 2 μm in depth. (Scale bar, 10 μm.)
Discussion
Two mutant plants, web1 and web2/pmi2, in which the chloroplast movement velocity is much slower than in WT plants, were isolated, and the corresponding disrupted genes were identified. We also found the cytosolic WEB1–PMI2 complex and genetic interactions of the WEB1 and PMI2 with JAC1. Blue light-induced reorganization of cp-actin filaments was regulated by both WEB1 and PMI2. These findings suggest that WEB1 and PMI2 form a cytosolic protein complex and cooperatively maintain the velocity of chloroplast movement via cp-actin filament regulation.
Although WEB1 was detected mostly in the soluble but not in the microsomal fraction under the protein extraction conditions used, GFP-WEB1 was predominantly localized to the cell periphery, and WEB1 self-association was detected only at the cell periphery by BiFC analysis. In this context, the WEB1 homomultimers might be localized in the soluble fraction close to the plasma membrane (e.g., the ectoplasm). Nevertheless, it is plausible that peripheral WEB1 homomultimers are not the active complex for chloroplast movement because the pmi2 mutant, which contained WEB1 homomultimers but not the WEB1–PMI2 complex, showed the same phenotype as the web1pmi2 double mutant. In contrast to WEB1, PMI2 self-association was not observed by BiFC assay, suggesting that PMI2 does not form homodimers. The definite function of WEB1 homomultimers at the cell periphery and further analysis for PMI2 self-association will be studied in the future.
We found that WEB1 and PMI2 physically interact in yeast and in planta and that the interaction occurred in the cytosol. Consistent with this result, CFP-WEB1 and YFP-PMI2 were colocalized in the cytosol, and WEB1 was a soluble protein. Because the web1 and pmi2 single mutants behaved similarly to the web1pmi2 double mutant, WEB1 is essential for PMI2 function and vice versa. Therefore, it is possible that cytosolic WEB1-PMI2 complex formation is necessary for normal chloroplast movement.
Analysis of chloroplast movement based on changes in leaf transmittance indicated that the web1 and pmi2 mutants were deficient only in the avoidance response and not in the accumulation response (13). However, the direct measurement of the velocity of chloroplast movement induced by irradiation with a blue light microbeam revealed that both the avoidance and the accumulation responses are disrupted in the web1 and pmi2 mutants. The difference in the results obtained from the direct (microbeam irradiation) and indirect (leaf transmittance) observations might have occurred because the light transmission measured by the indirect method only partly reflects the level of chloroplast movement.
The phenotype of the jac1 mutant suggested that JAC1 might function specifically in the accumulation response but not in the avoidance response pathway (11). Although web1 and pmi2 single mutants showed impaired avoidance responses, the jac1 mutation restored the avoidance response in the web1jac1 and pmi2jac1 double mutants to the level of the jac1 mutant. Thus, WEB1 and PMI2 promote the avoidance response by suppressing JAC1 activity, but they are not essential for the avoidance response itself; however, the mechanism underlying the weak accumulation response in web1 and pmi2 is still unknown. Under strong light conditions, signal transduction pathways for both the accumulation and the avoidance responses are activated, but the outcome is the avoidance response (1), suggesting that a mechanism for suppression or prevention of the accumulation response exists under strong light conditions so that the avoidance response is easily induced. Thus, it is plausible that WEB1 and PMI2 may be necessary to suppress or override the accumulation response pathway (i.e., JAC1 activity) under strong light conditions. Although a detailed mechanism, in which WEB1 and PMI2 suppress the JAC1 activity under strong light conditions, remains unknown, the genetic interaction and the Y2H results suggested that WEB1 and PMI2 suppress the JAC1 activity via unidentified factor to promote the avoidance response. Our previous work suggested that the JAC1 localizes to cytosol (11), like WEB1 and PMI2. Given that the three proteins colocalize in cytosol, the JAC1 suppression seems to occur in cytosol and the unidentified factor involved in the suppression is also likely cytosolic factor.
We revealed that blue light-induced cp-actin filament reorganization was impaired in both web1 and pmi2. The cp-actin filaments determine the velocity of chloroplast movement (9, 10), which suggests that the slow chloroplast movement in both web1 and pmi2 resulted from a defect in cp-actin filament regulation. Slow chloroplast movement was also observed in the kac1 single mutant, in which fewer cp-actin filaments were found compared with WT (10). Unlike the kac1 single mutant, the light-induced appearance of cp-actin filaments in both web1 and pmi2 was comparable to that of WT, but strong light-induced rearrangements and subsequent biased localization of cp-actin filaments were defective in these mutants, as in the phot2 mutant (9). Thus, WEB1 and PMI2 function in the phot2-mediated disappearance of cp-actin filaments rather than in the generation of cp-actin filaments (i.e., polymerization).
In the present study, we showed a possible pathway via protein–protein interactions for chloroplast velocity maintenance by cp-actin regulation. Our findings suggest that modification of the WEB1–PMI2 complex (or the pathway it mediates) improves chloroplast velocity for the adjustment of photosynthetic activity in plants.
Materials and Methods
Plant culture, the mutant screen, and map-based cloning were performed as described previously (4, 6, 11). Full-length cDNAs were isolated by the 5′ and 3′ rapid amplification of cDNA ends method, according to the manufacturer's instructions (Invitrogen). RT-PCR analysis was performed as described previously (11). Chloroplast photorelocation movement was analyzed by WBA in a growth chamber with a 3.6-kW xenon lamp (Ushio) and by the microbeam irradiation method with a custom-made system (24). Immunoblotting, fluorescent protein, and Y2H analyses are described in SI Materials and Methods. SI Materials and Methods includes full methods and any associated references.
Supplementary Material
Acknowledgments
We thank Nir Ohad (Tel-Aviv University, Tel-Aviv, Israel) for providing bimolecular fluorescence complementation vectors; Takatoshi Kagawa for technical advice regarding measurement of cytoplasmic motility; Hidenori Tsuboi for technical advice regarding microbeam irradiation; and Mineko Shimizu for technical assistance. This work was supported in part by Japanese Ministry of Education, Sports, Science, and Technology Grants 13139203 and 17084006 (to M.W.) and 21770050 (to S.-G.K); and Japan Society of Promotion of Science Grants 13304061, 16107002, and 20227001 (to M.W.) and 20870030 (to N.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007836107/-/DCSupplemental.
References
- 1.Suetsugu N, Wada M. Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol Chem. 2007;388:927–935. doi: 10.1515/BC.2007.118. [DOI] [PubMed] [Google Scholar]
- 2.Kasahara M, et al. Chloroplast avoidance movement reduces photodamage in plants. Nature. 2002;420:829–832. doi: 10.1038/nature01213. [DOI] [PubMed] [Google Scholar]
- 3.Sakai T, et al. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagawa T, et al. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 5.Jarillo JA, et al. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 6.Oikawa K, et al. Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oikawa K, et al. Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 2008;148:829–842. doi: 10.1104/pp.108.123075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt von Braun S, Schleiff E. The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta. 2008;227:1151–1159. doi: 10.1007/s00425-007-0688-7. [DOI] [PubMed] [Google Scholar]
- 9.Kadota A, et al. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:13106–13111. doi: 10.1073/pnas.0906250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suetsugu N, et al. Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:8860–8865. doi: 10.1073/pnas.0912773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suetsugu N, Kagawa T, Wada M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005;139:151–162. doi: 10.1104/pp.105.067371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBlasio SL, Luesse DL, Hangarter RP. A plant-specific protein essential for blue-light-induced chloroplast movements. Plant Physiol. 2005;139:101–114. doi: 10.1104/pp.105.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luesse DR, DeBlasio SL, Hangarter RP. Plastid movement impaired 2, a new gene involved in normal blue-light-induced chloroplast movements in Arabidopsis. Plant Physiol. 2006;141:1328–1337. doi: 10.1104/pp.106.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagawa T, Wada M. Velocity of chloroplast avoidance movement is fluence rate dependent. Photochem Photobiol Sci. 2004;3:592–595. doi: 10.1039/b316285k. [DOI] [PubMed] [Google Scholar]
- 15.Kagawa T, Wada M. Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol. 2000;41:84–93. doi: 10.1093/pcp/41.1.84. [DOI] [PubMed] [Google Scholar]
- 16.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu W, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 19.Bae W, et al. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol. 2008;10:220–227. doi: 10.1038/ncb1683. [DOI] [PubMed] [Google Scholar]
- 20.Grefen C, et al. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant. 2008;1:308–320. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]
- 21.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong SG, et al. Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 2006;45:994–1005. doi: 10.1111/j.1365-313X.2006.02667.x. [DOI] [PubMed] [Google Scholar]
- 24.Yatsuhashi H, Wada M. High-fluence rate responses in the light-oriented chloroplast movement in Adiantum protonemata. Plant Sci. 1990;68:87–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.