Abstract
The activation and recruitment of the small GTPase Rab7 to early endosome is a critical step for early to late endosome maturation, a process that requires the class III phosphatidylinositol 3-kinase (PI3KC3) and GTPase regulators. However, the molecular mechanism underlying Rab7 activation and endosome maturation is still poorly defined. Here we report that Rubicon, a component of the PI3KC3 complex, prevents endosome maturation through differential interactions with Rab7 and UVRAG. UVRAG activates PI3KC3 and C-VPS/HOPS, a guanine nucleotide exchange factor that catalyzes the exchange of GDP for GTP on Rab7. We demonstrate that Rubicon sequesters UVRAG from C-VPS/HOPS. Active GTP-bound Rab7 competes for Rubicon binding and releases UVRAG to associate with C-VPS/HOPS, which in turn promotes further loading of Rab7 with GTP. This feed-forward loop ensures rapid amplification of GTP-bound Rab7 and consequent stimulation of endosome maturation. Hence, Rubicon serves as a previously unknown Rab7 effector to ensure the proper progression of the endocytic pathway.
Keywords: autophagy, endocytosis, epidermal growth factor, autophagosome, Barkor/Atg14(L)
Endocytic transport involves the passage and sorting of cargo within different sets of vesicles that are marked by organelle-specific Rab GTPase proteins (1). In the endocytic pathway, early endosomes are characterized by the small GTPase Rab5, early endosomal antigen EEA1, and PI3KC3 (2–4). Rab5 regulates homotypic fusion of early endosomes. These early endosomes then mature into late endosomes, which will be targeted for lysosomal degradation or processing (5). Understanding the molecular mechanisms underlying endosome maturation is of clinical significance, as many bacteria (e.g., Mycobacterium tuberculosis) and viruses (e.g., HIV) interfere with phagolysosomal or endolysosomal maturation to avoid elimination in lysosomes (6).
The early-to-late endosome transition begins with the activation and recruitment of Rab7 to the subdomains of early endosomes bearing Rab5, followed by Rab5 displacement from the same vesicle (5, 7) or endosome fission with the formation of late-endosome-targeted transportation vesicles (8). The activation of Rab7 is required for endosome maturation. However, little is known about how Rab7 activation is controlled.
As a small GTPase, Rab7 regulates membrane trafficking by cycling between inactive (i.e., GDP-bound) and active (i.e., GTP-bound) conformations. The class C-VPS/HOPS (homotypic fusion and vacuole protein sorting) complex is the guanine nucleotide exchange factor (GEF) for Rab7 (9). The GEF promotes GDP-to-GTP transition, leading to Rab7 activation. Active GTP-bound Rab7 binds to Rab7 effectors and executes its function in vesicle tethering, docking, and fusion (10, 11). The GTPase activating proteins (GAP) inactivate Rab7, stimulating the conversion from the GTP-bound form to the GDP-bound form (12, 13). The C-VPS/HOPS complex also serves as a major downstream effector of active GTP-bound Rab7 (14). Recent evidence indicates that this complex is positively regulated by UVRAG, a component of PI3KC3 (15), which provides an interesting link between PI3KC3 and Rabs that are both crucial for endosome maturation.
We and others have recently purified a PI3KC3 holocomplex that includes hVPS34, p150, Beclin 1, UVRAG, and Barkor/Atg14(L) (16–20). PI3KC3 forms two mutually exclusive protein subcomplexes that localize to autophagosome or endosome and execute distinct functions. The autophagosomal complex is composed of the PI3KC3 core complex (hVPS34, p150, and Beclin 1) and Barkor/Atg14(L). Barkor/Atg14(L) is the targeting factor for this subcomplex to nascent autophagosomes (16–20). The endosomal complex consists of the PI3KC3 core complex and UVRAG. UVRAG positively regulates PI3KC3 activity and is required for autophagosome and endosome maturation probably via its direct interaction with C-VPS/HOPS (15, 21). The endosomal complex also contains Rubicon (Run domain protein as Beclin 1 interacting and cysteine-rich containing), which serves as a negative regulator of autophagosome maturation (18, 19). Although it has been suggested that Rubicon plays a negative role in endosome maturation, this function is still under debate.
Here we report that Rubicon is a key negative regulator in endosome maturation and that active Rab7 antagonizes this inhxibition. Rubicon is highly enriched on Rab5-positive early endosomes and sequesters UVRAG from C-VPS/HOPS. Active GTP-bound Rab7 competes for Rubicon binding and releases UVRAG. This release promotes the complex formation between UVRAG and C-VPS/HOPS to further activate Rab7. These events trigger and amplify the early-to-late endosome maturation.
Results
Rubicon Interacts with Rab7 via Its C Terminus.
We recently identified a Beclin 1/PI3KC3 complex using Beclin 1 as bait in human osteosarcoma U2OS cells (16). This complex was composed of the PI3K catalytic subunit hVps34, p150 regulatory subunit, UVRAG, and Barkor/Atg14(L). In the same complex, Rubicon (also called p120 or Baron) was also identified (16, 18–20). Rubicon contains 972 amino acids, with a recognizable RUN domain (named after RPIP8/UNC-14/NESCA) that is shared by a group of proteins interacting with small GTPases (22–24).
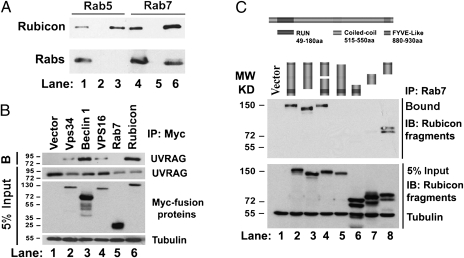
Given Rubicon's RUN domain and the importance of Rab5 and Rab7 in endosome maturation, we tested whether Rubicon interacts with the Rab family. Rab7, but not Rab5, coimmunoprecipitated with Rubicon (Fig. 1A). As Rubicon is known to associate with UVRAG in the PI3KC3 complex (18, 19), we investigated whether Rab7 coexists with UVRAG in the same protein complex. UVRAG coimmunoprecipitated with Myc-tagged hVps34, Beclin 1, Vps16 (a component of C-VPS/HOPS complex), Rab7, and Rubicon. UVRAG consistently interacted with the PI3KC3 components including Vps34, Beclin 1, and Rubicon (Fig. 1B). UVRAG also associated with Vps16 (Fig. 1B). However, no Rab7-UVRAG interaction could be detected in this coimmunoprecipitation assay. Therefore, Rab7 and UVRAG form distinct complexes with Rubicon.
Fig. 1.
Rubicon interacts with Rab7 via its C terminus. (A) Rubicon interacts with Rab7 but not Rab5. Rubicon-Flag vector was cotransfected with either Rab5-Myc (lanes 1–3) or Rab7-Myc vectors (lanes 4–6). The whole 293T cell lysates (lanes 1 and 4) were immunoprecipitated with anti-Flag antibody (lanes 3 and 6) or control IgG (lanes 2 and 5), followed by immunoblotting with anti-Flag antibody for Rubicon and anti-Myc antibody for Rabs. (B) Rubicon interacts with UVRAG and Rab7 in different complexes. Myc tagged Vps34, Beclin 1, Vps16, Rab7, and Rubicon were expressed in HEK293T cells and immunoprecipitated (IP) with anti-Myc antibody. The proteins indicated were detected by immunoblotting (IB) in the bound (B) and input fractions. (C) The CT domain of Rubicon is required for Rab7 binding. A vector expressing HA-tagged Rab7 was cotransfected with EGFP-Rubicon-Flag (WT) vector or the vectors expressing different mutants, followed by immunoprecipitation with anti-HA antibody (Rab7) and immunoblotting with the antibodies indicated.
We further refined the interaction domain between Rubicon and Rab7. A series of Rubicon deletion mutants were generated, including mutants lacking the RUN, FYVE-like, or coiled-coil domains (CCD) from full-length Rubicon. We also created three large polypeptide fragments containing the RUN, CCD, or FYVE-like domains (Fig. 1C). The interaction of these mutants with Rab7 was evaluated by Rab7 immunoprecipitation assay. Full-length Rubicon, rather than vector alone, interacted with Rab7 (Fig. 1C, lanes 1–2). The RUN and CCD domains were dispensable for Rab7 interaction (Fig. 1C, lanes 3–4 and 6–7), whereas a C-terminal (CT) region (aa 880–972) was required (Fig. 1C, lane 5). A larger C terminus region of Rubicon (aa 600–972) containing the CT domain was sufficient for Rubicon to interact with Rab7 (Fig. 1C, lane 8).
Rubicon Preferentially Binds to GTP-Bound Rab7.
Rab7 regulates membrane trafficking by cycling between inactive (i.e., GDP-bound) and active (i.e., GTP-bound) conformations (25, 26). We then tested whether the guanine nucleotide cycling of Rab7 is crucial for its interaction with Rubicon. Coimmunoprecipitation experiments in 293T cells showed Rubicon binding to WT Rab7. Rab7 exhibited a relatively stronger binding to a constitutively active GTP-bound Rab7 Q67L mutant protein and much weaker binding to a GDP-bound Rab7 T22N mutant protein (Fig. 2A). To further confirm that the GTP-loaded Rab7 binds to Rubicon, we performed a coimmunoprecipitation experiment in the presence of excess amount of nonhydrolyzable GTP (GTP-γ-S) or GDP (GDP-β-S). The loading of GDP (GDP-β-S), rather than GTP (GTP-γ-S), decreased the Rab7–Rubicon interaction (Fig. S1). These results suggest that Rubicon could preferentially bind to Rab7 in GTP-bound form rather than GDP-bound form.
Fig. 2.
Rubicon preferentially binds to GTP-bound Rab7. (A) Rubicon binds to GTP-bound Rab7 in vivo. Rubicon-Flag vector was cotransfected in HEK293T cells with Rab7-Myc vectors expressing Rab7-WT, GTP-bound Rab7 Q67L mutant, or GDP-bound Rab7 T22N mutant, followed by immunoprecipitation with anti-Myc antibody (Rab7) and immunoblotting with the antibodies indicated. (B) Rab7 differentially interacts with Rubicon and p150. Purified recombinant HA-tagged Rab7 (0.1 μM) was incubated with recombinant Flag-tagged p150, Vps34, Beclin 1, UVRAG, Barkor/Atg14(L), or Rubicon (0.1 μM) in an in vitro pull-down assay using HA affinity beads. The bound and input proteins were detected by anti-Flag or HA antibodies. (C) Rubicon directly binds GTP-bound Rab7. Recombinant full-length Flag-tagged p150, Vps34, or Rubicon (0.1 μM) were incubated with HA affinity beads prebound with recombinant HA-tagged Rab7 WT, T22N, or Q67L mutant proteins (0.1 μM). The bound proteins were resolved by 7.5% SDS/PAGE and analyzed by Western blot using anti-Flag antibody.
To test for a direct interaction between Rab7 and Rubicon in a nucleotide-dependent manner, we expressed and purified recombinant HA-tagged Rab7 WT, Q67L, and T22N mutants from Escherichia coli (Fig. S2A). We also expressed and purified recombinant full-length Flag-tagged Vps34, UVRAG, Beclin 1, Barkor/Atg14(L), and Rubicon from baculovirus-infected insect cells or mammalian cells (Fig. S2B), and evaluated their binding to Rab7. We then performed a pull-down assay with all purified components and observed that both p150 and Rubicon, but not other PI3KC3 components, bind directly to Rab7 (Fig. 2B). We further tested the binding of different forms of Rab7 proteins to p150 and Rubicon. The binding of p150 to WT Rab7 was clearly detected (Fig. 2C). p150 bound to GDP-bound Rab7 T22N much more weakly; essentially no binding of p150 to the GTP-bound Rab7 Q67L mutant was observed (Fig. 2C). This result is consistent with the published observation that p150 preferentially interacts with nucleotide-free Rab7 (27). In contrast to p150, Rubicon preferentially interacted with GTP-bound Rab7 (Fig. 2C), suggesting that it might function as a Rab7 effector via direct interaction.
Rab7 Competes with UVRAG for Rubicon Binding.
Rubicon interacts with UVRAG and Rab7 in distinct protein complexes (Fig. 1B). We speculated that Rab7, especially the GTP-bound form, might then compete with UVRAG for Rubicon binding. Thus, we overexpressed UVRAG to test its effect on Rubicon–Rab7 interaction. UVRAG overexpression dramatically decreased the amount of Rubicon coimmunoprecipitated with Rab7 or Rab7 pulled down by Rubicon (Fig. 3A). Conversely, Rab7 overexpression also compromised the UVRAG–Rubicon interaction in the immunoprecipitation assay (Fig. 3B). These results suggest that Rab7 and UVRAG compete for interaction with Rubicon in vivo. To test if Rab7 competes with UVRAG for Rubicon binding in a nucleotide-dependent manner, Rab7 WT, GDP-bound T22N mutant, or GTP-bound Q67L mutant proteins were expressed and tested for their effects on Rubicon–UVRAG interaction. The GTP-bound Rab7 Q67L mutant competed with UVRAG for Rubicon binding much more efficiently than the GDP-bound Rab7 T22N mutant (Fig. 3C). We therefore concluded that GTP-bound Rab7 competes with UVRAG for Rubicon binding in vivo.
Fig. 3.
GTP-bound Rab7 competes with UVRAG for Rubicon binding. (A) Overexpression of UVRAG compromises the Rab7–Rubicon interaction. Different concentrations of UVRAG were overexpressed in HEK293T cells (Top), and the resulting cell lysates were immunoprecipitated with anti-Rab7 antibody (Middle) or anti-Rubicon antibody (Bottom). The Rab7 or Rubicon immunocomplex was then probed for the proteins indicated. The normalized binding of Rubicon and Rab7 was quantified as the relative ratio of bound/input (B/I). The B/I in the control cells was set as 1.0. (B) Overexpression of Rab7 decreases UVRAG–Rubicon interaction. Rab7 was expressed in HEK293T cells. The resulting cell lysate was then immunoprecipitated with anti-Rubicon antibody. The bound and input fractions were probed for the proteins indicated. (C) GTP–Rab7 competes Rubicon from UVRAG in vivo. Different forms of Rab7 (WT, T22N, Q67L) were expressed and the resulting cell lysate was immunoprecipitated with anti-UVRAG antibody. The bound and input were probed for the proteins indicated. (D) Recombinant purified GST-tagged Rab7 WT and mutants from E. coli were analyzed by 10% SDS/PAGE followed by Coomassie blue staining. (E) In vitro competition among Rubicon, UVRAG, and different forms of Rab7. Rubicon-Flag-His (0.1 μM) was bound to Ni column first and then incubated with UVRAG-Flag (0.1 μM) to form a Rubicon–UVRAG complex. Different concentrations (0.1, 0.5, 2 μM) of Rab7 proteins were incubated with the Rubicon–UVRAG complex. The bound proteins were analyzed by Western blotting using anti-Flag and anti-Rab7 antibodies.
We further tested the direct competition between Rab7 and UVRAG for Rubicon binding in vitro. We expressed and purified recombinant full-length GST-tagged Rab7 WT, GDP-bound T22N mutant, and GTP-bound Q67L mutant proteins from E. coli (Fig. 3D) and incubated these proteins with the preassembled Rubicon–UVRAG complex in a purified system. Rab7 Q67L mutant competed with the UVRAG–Rubicon interaction with the highest efficiency, whereas the GDP-bound Rab7 T22N had almost no effect on this interaction (Fig. 3E). This assay bolsters our previous data by providing direct evidence that Rab7 competes for Rubicon from UVRAG in a nucleotide-dependent manner.
Rubicon Is Highly Enriched on Rab5-Decorated Early Endosomes.
To explore the potential role of Rubicon in endosome organization, we investigated its subcellular localization in human osteosarcoma U2OS cells stably expressing Flag-Rubicon. We generated a cell line that inducibly expresses Flag-Rubicon under the control of doxycycline. When Flag-Rubicon expression was induced by an extremely low dose of doxycycline (1 ng/mL), Flag-Rubicon was detected at a level comparable to endogenous Rubicon (Fig. S3A). Flag-Rubicon expressed at physiological level displayed a significant overlap with early endosome markers EEA1, UVRAG, and FYVE2 (Fig. S3B). We then tested Rubicon colocalization with Rab7 in Rubicon-expressing cells. Only limited colocalization between Rubicon and Rab7 could be observed in cells expressing Rubicon at a physiological level. No colocalization was detected between Rubicon and an inactive Rab7 T22N GDP-bound form (Fig. S3C). Rab7 colocalization with Rubicon was only apparent when a constitutively active GTP-bound Rab7 Q67L form was expressed (Fig. S3C) or Rubicon was expressed at a high level (Fig. S3D). This observation agrees with our in vitro binding results that Rubicon preferentially binds to active GTP-bound Rab7.
We next compared the localization of Rubicon expressed at a physiological level with Rab5 and two Rab5 mutants, GDP-bound S34N and GTP-bound Q79L. In cells costained for Rubicon and Rab5 (Fig. 4A, Top), Rubicon significantly overlapped with Rab5 in large cellular puncta. Most Rubicon signal colocalized or was detected adjacent to Rab5. However, Rubicon puncta did not overlap with the GDP-bound S34N form of Rab5 (Fig. 4A, Middle). Expression of the Rab5 GTP-bound Q79L mutant stimulates homotypic fusion of early endosomes and leads to formation of large ring-like membrane structures (28). These enlarged endosomes are considered arrested at the convergent point of endosome maturation after acquiring Rab7, as Rab5 cannot be displaced as a result of inefficient GTP hydrolysis (5, 8, 29). In Rab5 Q79L-expressing cells, Rubicon was highly enriched in subdomains of large ring-like, Rab5-positive early endosomal structures (Fig. 4A, Bottom). These data showed that Rubicon localizes to enlarged early endosomes and subdomains of maturing endosomes.
Fig. 4.
Rubicon localizes to maturing early endosomes. (A) Rubicon localizes to maturing early endosomes. U2OS cells expressing Rubicon-Flag were transfected with GFP-Rab5-WT, GFP-Rab5-S34N, or GFP-Rab5-Q79L and observed under a fluorescence microscope. Cy3-conjugated M2 antibody was used to label Rubicon-Flag. (Scale bar: 5 μm.) Framed areas of Rab5 Q79L-expressing cells are enlarged (Bottom). (B) Rab7 GTP-bound form disrupts the overlap of Rubicon–UVRAG. Mock transfection: GDP-bound Rab7 T22N or GTP-bound Rab7 Q67L were coexpressed with GFP–UVRAG and Rubicon-Flag in U2OS cells. The subcellular localization of UVRAG (green) and Rubicon (red) was observed under a fluorescence microscope. (C) The number of cellular puncta positive for UVRAG and Rubicon was counted and plotted in cells described in B.
To confirm that Rab7 competes with UVRAG for Rubicon binding in vivo, we expressed GTP-bound Rab7 Q67L or GDP-bound Rab7 T22N and scored their effect on the colocalization of Rubicon and UVRAG. In the control mock-transfected cells, Rubicon colocalized well with UVRAG (Fig. 4B, Top, and 4C). The colocalization of these two proteins were not altered upon GDP-bound Rab7 T22N expression (Fig. 4B, Middle, and 4C), but significantly reduced upon GTP-bound Rab7 Q67L expression (Fig. 4B, Bottom, and 4C). This further supports our hypothesis that Rab7 GTP-bound form competes for Rubicon binding with UVRAG.
Rubicon Sequesters UVRAG from C-VPS/HOPS and Blocks Rab7 Activation.
UVRAG interacts with C-VPS/HOPS and activates its GEF activity, resulting in Rab7 activation (15). We speculated that Rubicon sequesters UVRAG from C-VPS/HOPS and blocks Rab7 activation. We tested the UVRAG-Vps16 (a C-VPS/HOPS component) interaction in Rubicon RNAi-depleted cells. The interaction between UVRAG and Vps16 was significantly increased in the absence of Rubicon (Fig. 5A). Conversely, in Rubicon-overexpressing cells, much less Vps16 coprecipitated with UVRAG (Fig. 5B). This demonstrates that Rubicon sequesters UVRAG from C-VPS/HOPS interaction.
Fig. 5.
Rubicon sequesters UVRAG from C-VPS/HOPS and negatively regulates Rab7 activation. (A) Rubicon depletion promotes the interaction between UVRAG and C-VPS/HOPS. Cell lysates from Rubicon WT or knockdown (KD) U2OS cells were immunoprecipitated with anti-UVRAG antibody. The indicated proteins were detected. (B) Rubicon overexpression decreases the binding of UVRAG to C-VPS/HOPS. HEK293T cells were transfected with Rubicon-Myc or vector alone. The cell lysates were immunoprecipitated with anti-UVRAG antibody. The indicated proteins were detected. (C) Depletion of Rubicon stimulates Rab7-RILP interaction. In cell lysates prepared from WT or Rubicon RNAi-depleted cells, Rab7 was immunoprecipitated and the resulting immunocomplexes were probed for Rab7 and RILP. 3-MA (10 mM) was used to treat both Rubicon WT and depleted cells for 8 h before collection. (D) Depletion of Rubicon promotes Rab7–Vps41 interaction. In cell lysates prepared from WT or Rubicon RNAi-depleted cells, Rab7 was immunoprecipitated and the resulting immunocomplex was probed for Rab7 and Vps41.
We hypothesize that Rubicon sequesters UVRAG from C-VPS/HOPS interaction to block Rab7 activation. If this is true, Rubicon depletion should lead to Rab7 activation, which could be detected by the enhanced interaction between the active GTP-bound Rab7 and Rab7 effectors. To test this hypothesis, we examined the interaction between Rab7 and a well known Rab7 effector RILP (Rab-interacting lysosomal protein) (30). RILP specifically interacts with GTP-bound Rab7; the amount of RILP coprecipitated with Rab7 could reflect the GTP-binding and activation of Rab7. In Rubicon-depleted cells, the association between Rab7 and RILP was dramatically increased (Fig. 5C), suggesting that Rab7 is activated. However, it is also possible that the loss of Rubicon frees up more GTP-bound Rab7 for RILP binding rather than Rab7 activation. To test this possibility, we expressed a constitutively active GTP bound Rab7 Q67L mutant in WT and Rubicon-depleted cells. Rab7 Q67L strongly bound to RILP but no further increase could be observed in Rubicon-depleted cells, whereas the Rab7–RILP interaction was clearly increased in the absence of Rubicon (Fig. S4A), further supporting the notion that the generation of GTP-bound Rab7 is probably up-regulated without Rubicon.
The interaction between Rab7 and another Rab7 effector, Vps41 (14), was also investigated. Consistently, the interaction between Rab7 and Vps41 was dramatically increased in Rubicon-depleted cells (Fig. 5D). The increased interactions of Rab7–Vps41 and UVRAG–Vps16 in Rubicon-depleted cells could be efficiently suppressed by the complementation of RNAi-resistant Rubicon (Fig. S4B), excluding the off-target effect of RNA interference. Furthermore, overexpression of Rubicon led to a reduced Vps41–Rab7 interaction (Fig. S5). Thus, Rubicon suppresses Rab7 activation through C-VPS/HOPS, illustrated by the increased interaction between Rab7 and Rab7 effectors in Rubicon-deficient cells. This conclusion is further supported by the translocation of Rab7 from perinuclear regions to cytosolic puncta in Rubicon-depleted cells (Fig. S6).
As UVRAG is a component of PI3KC3 through its interaction with Beclin 1 (21), we also investigated if Rubicon sequesters UVRAG from Beclin 1. Interestingly, the interaction between UVRAG and Beclin 1 or Vps34 was largely unaffected in the presence of Rab7 or Rubicon overexpression (Fig. S7), suggesting no competition between UVRAG–Beclin 1 interaction and UVRAG–Rubicon interaction, which is consistent with coexistence of these three proteins in the same protein complex (16, 20). To test if Rab7 activation in Rubicon-depleted cells is dependent on PI3KC3 activity, Rubicon-depleted cells were treated with a PI3KC3 inhibitor, 3-methyladenine (3-MA). The 3-MA treatment had no effect on Rab7–RILP interaction (Fig. 5C), suggesting that Rab7 is activated in a PI3KC3-independent manner.
Rubicon Controls Endosome Maturation and Endocytic Degradation.
Rubicon sequesters UVRAG from C-VPS/HOPS binding and blocks Rab7 activation. Given the essential role of Rab7 in endosome maturation, we therefore anticipated a negative role of Rubicon in the endocytic degradation pathway. We monitored the endocytic degradation of epidermal growth factor (EGF) receptor (EGFR) in the Rubicon-depleted cells. EGF was used to stimulate EGFR activation and subsequent degradation. In Rubicon-depleted cells, EGFR degradation was accelerated compared with what is observed in Rubicon WT cells (Fig. 6 A and B). EGFR degradation was also tested in Rubicon-overexpressing cells. EGFR turnover was significantly slowed in Rubicon-overexpressing cells (Fig. 6 C and D). These data further confirm the negative regulation of EGFR degradation by Rubicon.
Fig. 6.
Depletion of Rubicon alters EGFR degradation via endocytic pathway. (A) EGFR degradation is accelerated in Rubicon-depleted cells. EGFR degradation was examined in Rubicon RNAi-depleted 293T cells. Cells were serum-starved in DMEM-only medium for 10 h, followed by 200 ng/mL EGF addition to initiate EGFR degradation. Cells were collected at indicated time points and tested for EGFR level by Western Blotting. (B, D, and F) EGFR levels in A, C, and E were quantified by a Phosphorimager and normalized by calculating as the percentage of the initial receptor content. (C) EGFR degradation was tested in Rubicon overexpression cells. (E) EGFR degradation was examined in Rubicon-depleted 293T cells expressing vector alone, Rubicon WT, or Rubicon-ΔCT. (G) EGFR accumulates in large vacuoles in Rubicon-overexpressing cells. In U2OS cells expressing vector alone or Rubicon, the endogenous EGFR localization was detected by immunostaining using anti-EGFR antibody. (Scale bar: 5 μm.) (H) Number of vacuoles greater than 800 nm in diameter was counted and plotted in cells (G).
We complemented Rubicon-knockdown cells with various RNAi-resistant cDNAs for Rubicon WT or mutant forms and monitored EGFR degradation. WT Rubicon was able to rescue the accelerated EGFR degradation, whereas vector alone did not (Fig. 6 E and F). Importantly, the Rubicon CT deletion mutant that disrupts the interaction with Rab7 (Fig. 1C) failed to complement Rubicon defects (Fig. 6 E and F). Additionally, in Rubicon overexpression cells, EGFR was accumulated in the large vacuoles (Fig. 6 G and H). These results demonstrate that the Rab7 interaction is indispensable for Rubicon's function in the endocytic degradation.
Discussion
In this study, we have demonstrated a critical function for Rubicon in endocytic trafficking, which is to control the endosome maturation by regulating Rab7 and UVRAG. Several lines of evidence support this conclusion. First, Rubicon interacts with Rab7 and UVRAG in a mutually exclusive manner. Rubicon binds to UVRAG and sequesters it from the C-VPS/HOPS complex. Furthermore, GTP-bound Rab7 selectively interacts with Rubicon and competes with UVRAG–Rubicon binding, which in turn activates C-VPS/HOPS activity on Rab7. Finally, depletion of Rubicon promotes Rab7 activation and the endocytic degradation of EGFR. Hence, Rubicon negatively regulates Rab7 activation and endosome maturation as a Rab7 effector (Fig. S8).
We have established that GTP-bound Rab7 releases UVRAG from Rubicon sequestration. There are two consequences of this UVRAG release (Fig. S6). First, the released UVRAG binds and activates C-VPS/HOPS (15). Additionally, PI3KC3 activity may be affected, given that UVRAG is shown to positively regulate PI3KC3 (21). If that were the case, we would expect that active Rab7 could stimulate PI3KC3 activity. Interestingly, overexpressed Rab7 was able to stimulate PI3KC3 and partially compensate the PI3KC3 deficiency (27), supporting this idea. It is suggested that p150 interacts with Rab7 to fulfill this function. However, p150 preferentially binds to an inactive nucleotide free form rather than active GTP-bound form of Rab7 (27), which we also confirmed (Fig. 2C). This observation makes it difficult to explain the stimulation effect of Rab7 on PI3KC3 activation. Our study provides an alternative model in which GTP-bound Rab7 abolishes the Rubicon's sequestration of UVRAG and allows activation of PI3KC3 by UVRAG (Fig. S8).
UVRAG and Rubicon play critical functions not only in endosome maturation but also in autophagosome maturation (18, 19). Emerging evidence suggests that endocytic proteins are also involved in autophagy regulation (31, 32). Upon autophagosome formation, they fuse with endosomes to form amphisomes that eventually fuse with lysosomes or directly fuse with lysosomes to form autolysosomes. In hepatocytes, there are more than five times as many amphisomes formed as autolysosomes (33), suggesting that fusion with the endocytic vesicles could constitute a major part of autophagosome maturation, at least in certain cell types. Endosomes have been implicated in autophagosome maturation as a result of the involvement of ESCRTIII (34, 35) and COPI (36) in autophagy regulation. In this study, we have provided a molecular linkage connecting PI3KC3, autophagy, and endocytosis, in which Rubicon serves as a key negative regulator.
In summary, Rubicon serves as a Rab7 effector that negatively regulates endosome maturation. Dissection of this molecular switch will provide important insight into the mechanisms underlying the endocytic and autophagic pathways. Such information should be critical for the development of novel therapeutic tools for multiple human pathological conditions that result from the dysfunction of autophagic and endocytic degradation pathways.
Materials and Methods
Cell Culture, Plasmids, and Antibodies.
Tetracycline-inducible cell lines were established, and immunoblotting and immunoprecipitation were performed as described (16, 37). The full-length cDNA for human Rubicon (KIAA0226) was generated by PCRs on the basis of a partial cDNA clone purchased from Kazusa. Additional information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jae Jung (University of Southern California, Los Angeles, CA) for UVRAG constructs, Randy Schekman for discussion, Suzanne Pfeffer for suggestions, and Livia Wilz, Mary Grace Lin, and Jing Zhang for the critical reading of the manuscript. This work was supported by a New Investigator Award for Aging from the Ellison Medical Foundation, the Hellman Family Fund, and National Institutes of Health Grant R01 CA133228 (to Q.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010554107/-/DCSupplemental.
References
- 1.Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci USA. 2009;106:14185–14186. doi: 10.1073/pnas.0907725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spang A. On the fate of early endosomes. Biol Chem. 2009;390:753–759. doi: 10.1515/BC.2009.056. [DOI] [PubMed] [Google Scholar]
- 3.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 4.Wickner W, Haas A. Yeast homotypic vacuole fusion: A window on organelle trafficking mechanisms. Annu Rev Biochem. 2000;69:247–275. doi: 10.1146/annurev.biochem.69.1.247. [DOI] [PubMed] [Google Scholar]
- 5.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, et al. Endosomal membrane traffic: convergence point targeted by Mycobacterium tuberculosis and HIV. Cell Microbiol. 2004;6:999–1009. doi: 10.1111/j.1462-5822.2004.00449.x. [DOI] [PubMed] [Google Scholar]
- 7.Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markgraf DF, Peplowska K, Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581:2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 11.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XM, Walsh B, Mitchell CA, Rowe T. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem Biophys Res Commun. 2005;335:154–161. doi: 10.1016/j.bbrc.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 13.Frasa MA, et al. Armus is a Rac1 effector that inactivates Rab7 and regulates E-cadherin degradation. Curr Biol. 2010;20:198–208. doi: 10.1016/j.cub.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 14.Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 22.Mari M, Macia E, Le Marchand-Brustel Y, Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J Biol Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- 23.Kukimoto-Niino M, et al. Crystal structure of the RUN domain of the RAP2-interacting protein x. J Biol Chem. 2006;281:31843–31853. doi: 10.1074/jbc.M604960200. [DOI] [PubMed] [Google Scholar]
- 24.Recacha R, et al. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- 26.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 27.Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 28.Bucci C, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 29.Stenmark H, et al. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 33.Strømhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 36.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan W, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.