Abstract
The tumor suppressor p53 is a master sensor of stress, and posttranslational modifications are key in controlling its stability and transcriptional activities. p53 can be phosphorylated on at least 23 Ser/Thr residues, the majority of which are phosphorylated by stress-related kinases. An exception is Ser315 in human p53 (Ser312 in mouse), which is predominantly phosphorylated by cell cycle-related kinases. To understand the biological importance of Ser312 phosphorylation in vivo, we generated p53Ser312Ala knock-in mice. We show here that, although Ser312 is not essential for mouse life span under normal physiological conditions, Ser312Ala mutation dampens p53’s activity during embryonic development. This is evident from its partial rescue of embryonic lethality caused by Mdm4 deletion. In agreement with the notion that Ser312 mutation weakens p53 function, Ser312Ala mice are also more susceptible to tumorigenesis following a sublethal ionizing radiation dose. Importantly, in the cohort studied, Ser312 mutation predisposes mice to develop thymic lymphomas and liver tumors, partly due to p53Ser312Ala's inability to fully induce a set of p53 target genes including p21 and cyclin G1. Thus, we demonstrate that phosphorylation of Ser312 is required for p53 to function fully as a tumor suppressor in vivo.
Keywords: Mdm4, lymphoma, liver, knock-in, mouse
One of the best studied tumor suppressor genes is p53. The importance of its tumor suppressive role is illustrated in humans by the congenital Li-Fraumeni syndrome, where a mutation in the p53 gene results in the development of a range of malignancies, and the observation that the p53 gene undergoes inactivating mutations in around 50% of human tumors. p53 is normally present in the cell at low levels but increases in abundance in response to cellular stresses such as DNA damage, largely due to inhibition of its proteasomal degradation. Once elevated, it prevents potentially harmful damage being incurred following cell division, either by stalling the cell cycle until the damage is repaired or by preventing division entirely by inducing apoptosis or senescence. This is achieved largely by its ability to act as a transcription factor and by its regulation of target genes involved in various biological functions.
p53 is subject to many posttranslational modifications, such as acetylation and phosphorylation, which seem to both stabilize p53 and activate its transcriptional activity (1, 2). p53 is phosphorylated at 23 sites, mostly found within the N- and C-terminal domains that form the molecule's regulatory regions. Most of these serine and threonine residues are phosphorylated by stress-related kinases such as ATM/ATR and Chk2. However, Ser315 of human p53 (Ser312 in mouse) is phosphorylated by kinases involved in cell cycle progression, such as Aurora kinase A and the cyclin-dependent kinases cdk2 and cdk9 (3–6). Ser315 phosphorylation is required for the E2F family to enhance p53-mediated transcription and apoptosis (7) and is one of the sites phosphorylated when it interacts with the peptidyl-prolyl isomerase Pin1 following DNA damage to alter its conformation and increase its stability and transactivation activity (8, 9). Although phosphorylation at Ser315 in these instances seems to elevate p53 stability and positively contribute to tumor suppression, its phosphorylation by Aurora kinase A (involved in chromosome segregation and function) results in its destabilization (5). Following endoplasmic reticulum stress, phosphorylation of Ser315 alongside Ser376 results in p53’s relocation to the cytoplasm, inhibiting p53-mediated apoptosis (10). This suggests that Ser315 phosphorylation may obstruct p53’s role in tumor suppression.
To establish a physiological role for the phosphorylation of p53 at Ser315, we generated mice possessing a Ser → Ala mutation at codon 312 (equivalent to human codon 315). p53Ser312Ala mice were born normally and displayed no overt phenotype. However, following a sublethal dose of ionizing radiation (IR), they were more susceptible to tumor development, particularly in the thymus and liver. We conclude that Ser312 phosphorylation contributes to tumor suppression by p53 in response to ionizing radiation in vivo.
Results
Phosphorylation of p53 at Ser312 Does Not Affect Mouse Life Span.
Ser312 in murine p53 is equivalent to Ser315 in human p53 (Fig. 1A). To study the physiological effects of phosphorylating p53 at Ser312, knock-in mice were generated carrying a TCT → GCC (Ser → Ala) mutation in codon 312 of the mouse p53 gene (Fig. S1). The mutation was identified by PCR using primers specific for either the wild-type (WT) or mutant allele. p53 cDNA was isolated by RT-PCR from p53WT, heterozygous (p53312A/+) and mutant (p53312A/A) littermates, and the mutation was confirmed by sequencing (Fig. 1B). The entire p53 ORF was sequenced to confirm that no other mutations were present.
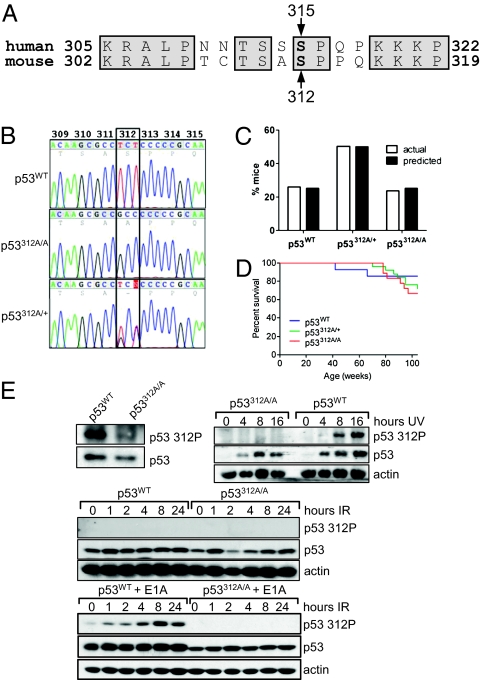
Fig. 1.
Generation of p53312A/A knock-in mice and verification of mutation. (A) Alignment of partial amino acid sequences from human and mouse p53, indicating homology (shaded boxes) between human Ser315 and mouse Ser312. (B) Chromatogram showing the presence of the TCT → GCC mutation at codon 312 in p53312A/+ heterozygous and p53312A/A homozygous mice. p53 cDNA was reverse-transcribed from RNA isolated from embryos of the indicated genotype. (C) p53312A/A mice are born at the predicted frequency and (D) have a normal life span compared with p53WT and p53312A/+ mice. (E, Upper Left) Immunoblot demonstrating lack of phosphorylation of p53 Ser312 in mutant MEFs. Total p53 was immunoprecipitated from untreated MEFs, and anti-p53 phospho-ser312 was used to show phosphorylation of this residue in p53WT, but not in p53312A/A, cells. MEFs were exposed to either UV (10 Jm−2) (Upper Right) or IR (4 Gy) (Middle), and p53 levels and phosphorylation status were determined at the indicated times by immunoblot. (Lower) MEFs were transformed with E1A, exposed to IR (4 Gy), and harvested at the indicated times.
Mice homozygous for Ala at codon 312 (p53312A/A mice) were viable, displayed no obvious developmental abnormalities, were born at the expected frequency (Fig. 1C), and survived to maturity. Cohorts of 11 p53WT, 25 p53312A/+, and 18 p53312A/A mice with a similar gender distribution for each genotype were followed over a period of 2 y to determine whether the mutation had a detrimental effect on life span. The p53312A/A mice survived for a time period comparable to the p53WT and p53312A/+ mice and did not develop spontaneous tumors at a significantly higher rate (P > 0.05) (Fig. 1D).
To verify that phosphorylation was not occurring in the p53312A/A mice, an antibody was raised against the phosphorylated Ser312 residue in WT p53. Using this antibody, Ser312 was phosphorylated in p53WT mouse embryonic fibroblasts (MEFs) but not in p53312A/A MEFs following immunoprecipitation with an antibody to total p53 (Fig. 1E, top left). As Ser312 has been reported to be phosphorylated in response to UV (11), UV-irradiated MEFs were analyzed with the anti-phosphoserine 312 antibody. Although Ser312 phosphorylation is undetecteable in nonirradiated MEFs, Ser312 is phosphorylated 8h after UV irradiation in WT, but not p53312A/A cells, confirming the antibody's specificity (Fig. 1E, Top Right). Next, we determined whether Ser312 is phosphorylated following IR. Phosphorylation of p53 at Ser312 could not be detected using this antibody after IR (Fig. 1E, Middle). As MEFs are primary cells, we transformed them with adenovirus E1A and analyzed Ser312 phosphorylation after IR in cells undergoing oncogenic stress. Ser312 was able to be phosphorylated in p53WT E1A-transformed cells, although not in p53312A/A cells (Fig. 1E, Bottom). We measured the ability of p53WT versus p53312A/A to influence the ability of E1A to immortalize MEFs. The difference was very small and not statistically significant, which agrees with the subtle effect of the mutation (Fig. S2).
p53312A/A Partially Rescues Embryonic Lethality Caused by Loss of Mdm4.
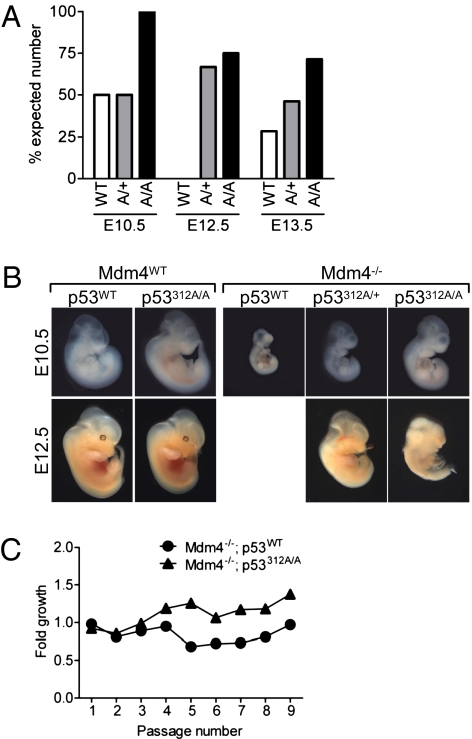
To investigate whether Ser312 could affect the activity of p53, we used a system in which p53 activity was elevated. In mice, the targeted deletion of Mdm4, an inhibitor of p53, results in increased p53 activity and embryonic lethality. This embryonic lethality can be entirely overcome by the simultaneous deletion of p53 (12–14). To investigate whether the Ser312Ala mutation could affect p53 activity in vivo, we isolated embryos from intercrosses between Mdm4+/−p53312A/+ mice. The Mdm4 deletion was caused by a gene trap insertion in the Mdm4 gene, 194 bp upstream of exon 2 (12). In an Mdm4-null background at embryonic day 10.5 (E10.5), only 50% of embryos were obtained with WT p53 or p53312A/+ compared with the expected Mendelian ratios (Fig. 2A and Table 1). The embryos isolated with genotypes of either Mdm4−/−p53WT or Mdm4−/−p53312A/+ were grossly abnormal when compared with WT embryos (Fig. 2B). However, the number of Mdm4−/−p53312A/A embryos obtained at E10.5 was 100% of the predicted number based on Mendelian segregation, suggesting that Ser312Ala mutation may reduce p53’s activity.
Fig. 2.
p53312A/A mice are able to partially rescue Mdm4−/− embryonic lethality. (A) Survival of Mdm4−/− embryos relative to the expected Mendelian frequency. (B) WT, Mdm4+/+p53312A/A, Mdm4−/−p53WT, Mdm4−/−p53312A/+, and Mdm4−/−p53312A/A embryos at embryonic day 10.5 (E10.5) and E12.5. No Mdm4−/− embryos with WT p53 were isolated at E12.5. (C) Fold growth of Mdm4−/− MEFs following serial passage.
Table 1.
Partial rescue of Mdm4−/− embryonic lethality by p53312A/A
| p53 | E10.5 | E12.5 | E13.5 | E14.5 | |
| Mdm4WT | WT | 4 (4) | 7 (4) | 9 (7) | 5 (4) |
| 312 A/+ | 12 (8) | 21 (9) | 16 (13) | 8 (8) | |
| 312 A/A | 6 (4) | 13 (4) | 4 (7) | 5 (4) | |
| Mdm4+/− | WT | 5 (8) | 9 (9) | 16 (13) | 10 (8) |
| 312 A/+ | 19 (15) | 8 (18) | 37 (27) | 21 (16) | |
| 312 A/A | 5 (8) | 2 (9) | 11 (13) | 12 (8) | |
| Mdm4−/− | WT | 2 (4) | 0 (4) | 2 (7) | 0 (4) |
| 312 A/+ | 4 (8) | 6 (9) | 6 (13) | 3 (8) | |
| 312 A/A | 4 (4) | 3 (4) | 5 (7) | 0 (4) | |
| Total | 61 | 69 | 106 | 64 |
The number of embryos identified at increasing stages of development following a cross of Mdm4+/−p53312A/+ mice. The numbers in parentheses are the predicted numbers based on Mendelian segregation. E, embryonic day.
This hypothesis was supported by the frequency of embryos recovered at later stages of development (up to E13.5). Sixty-nine embryos were examined at E12.5 and, as expected, none of them carried a Mdm4−/−p53WT genotype. We obtained a reduced number of Mdm4−/−p53312A/+ embryos (66%) (Fig. 2A and Table 1), and 75% of the predicted number for Mdm4−/−p53312A/A. At E12.5, although some of the embryos possessed severe growth defects, others (4/6 for Mdm4−/−p53312A/+) had developed further, as shown in Fig. 2B. A similar trend was also seen in embryos collected at E13.5 (Fig. 2A and Table 1). In a cohort of 106, we unexpectedly obtained 2 Mdm4−/−p53WT embryos.
Embryos isolated at E13.5 were used to make MEFs. Of the two Mdm4−/−p53WT embryos isolated at this stage, only one produced cells that grew, albeit slowly (Fig. 2C). Similarly, although six Mdm4−/−p53312A/+ embryos were identified, only one produced viable cells. However, fibroblasts from 3/5 Mdm4−/−p53312A/A embryos were able to proliferate in culture (Fig. 2C). Very few Mdm4-null embryos were obtained at E14.5 irrespective of p53 status (Table 1). Therefore, p53Ser312Ala mutation in one or both alleles can partially protect against embryonic lethality caused by Mdm4 loss, suggesting that Ser312 phosphorylation is necessary for p53 to be fully active.
Tumor Predisposition in p53312A/A Mice Exposed to Ionizing Irradiation.
To discover whether p53312A/A mice are more susceptible to tumor formation following exposure to a carcinogenic inducer, we exposed 3-wk-old mice to a single 4-Gy dose of IR. The study group consisted of 16 p53WT (8 females and 8 males), 21 p53312A/+ (10 females and 11 males), and 22 p53312A/A (12 females and 10 males) mice. This cohort had a mixed 129SvEv/C57BL6/J/ FVB background and so had black, agouti, or white coats (10 agouti, 21 black, and 28 white) (Table S1).
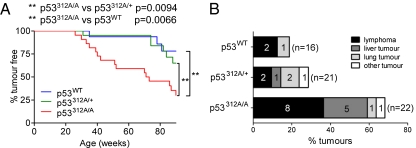
p53312A/A mice began to develop tumors at ≈25 wk. By 52 wk, 9/22 p53312A/A mice (40.9% of the cohort) had developed tumors, compared with 1/21 (4.8%) heterozygous and 1/16 (6.3%) p53WT mice. Among a group of age-matched mice not exposed to IR, none developed tumors during this time period. No tumors were observed in mice of any genotype between 52 and 70 wk. After 70 wk, the mice began to develop a variety of tumors. When overall tumor incidence was assessed after 90 wk, p53312A/A mice were significantly more susceptible to tumor development following IR exposure than their p53WT or p53312A/+ counterparts (p53312A/A vs. p53312A/+, P = 0.0094; p53312A/A vs. p53WT, P = 0.0066) (Fig. 3A). The types of tumors observed included lymphomas, sarcomas, and solid tumors in liver and lung (Fig. 3B). The experiment was terminated at 90 wk post irradiation, and the remaining mice were tumor-free at the end of the study.
Fig. 3.
p53 Ser312Ala mutation predisposes mice to tumor development following IR. (A) Survival curve showing overall tumor incidence over the course of the study. (B) The incidence of different tumor types by genotype. p53312A/A mice were significantly more prone to lymphomas and liver tumors. The incidence of lung tumors (the other main tumor group found during this study) was not significant.
The increased tumor susceptibility of p53312A/A mice in response to IR implies that phosphorylation of murine p53 at Ser312 contributes to p53’s tumor suppression in response to IR. We observed biphasic tumor onsets (25–52 wk and 70–90 wk) in this study, with an intervening 18-wk tumor-free period. Importantly, p53312A/A mice were significantly more prone to tumor development than p53WT or p53312A/+ mice in both phases.
Mice Carrying the Ser312Ala Mutation Are Initially Susceptible to Lymphoma Development.
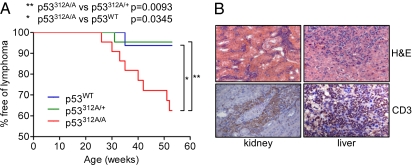
The tumors that developed within the first phase of tumor onset (25–52 wk) were almost all lymphomas, the exception being a sarcoma found in the intestinal wall of a p53312A/A mouse. The lymphomas found between 26 and 40 wk of age were all thymic lymphomas (n = 5; 22.7% of all p53312A/A mice). In contrast, lymphomas that arose between 42 and 52 wk (n = 3; 13.6%) were generalized lymphomas that had disseminated to the liver and kidney (Fig. 4B). The affected tissues from the three mice with generalized lymphomas were stained for B- and T-cell markers to determine their origin. One stained positively for the T-cell marker CD3 (Fig. 4B), whereas the other mice did not show positive staining for either CD3 or the B-cell marker B220. Disregarding the sarcoma, p53312A/A mice were significantly more susceptible to lymphoma development (p53312A/A vs. p53312A/+, P = 0.0093; p53312A/A vs. p53WT, P = 0.0345) (Fig. 4A). All of the p53312A/A mice that developed lymphomas (n = 8) had a white coat. Five of the eight mice were female, indicating that there was no gender bias in lymphoma development in this cohort. The majority of p53312A/+ and p53WT mice with white coats were tumor-free at this stage of the study. These data demonstrate that p53312A/A mice are predisposed to lymphomas and that their genetic background may influence tumor development.
Fig. 4.
p53312A/A mice are more susceptible to lymphoma development following exposure to IR. (A) Survival curve showing the occurrence of lymphomas in the first year following exposure to 4 Gy IR. Up to 40 wk, all lymphomas were found in the thymus (n = 5); later lymphomas (n = 3) were generalized lymphomas that had spread to the liver and kidney. (B) CD3 staining of liver and kidney from a generalized lymphoma.
As a link has been made between the development of thymic lymphomas in p53-null mice and the resistance of their thymocytes to DNA damage-induced apoptosis (15–18), we investigated whether the Ser312Ala mutation could compromise p53’s induction of apoptosis in response to IR. Thymocytes from p53WT and p53312A/A mice were exposed to a variety of apoptotic stimuli, including IR, and apoptosis was assessed by Annexin V binding (Fig. S3). Little difference was detected in the levels of apoptosis induced by any of the stimuli. Therefore, apoptotic failure is not behind the increased incidence of thymic lymphomas seen in p53312A/A mice.
Irradiated p53312A/A Mice Are Susceptible to Liver Hyperplasia at a Later Stage.
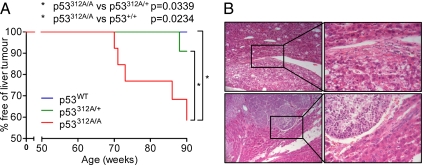
Following the occurrence of the lymphomas, no tumors were observed in mice of any genotype for a period of 18 wk. After 70 wk, however, the mice began to develop various tumors, most notably liver tumors (predominantly in p53312A/A mice). Of the surviving 13 p53312A/A mice, 5 (38.5% of remaining p53312A/A mice, 22.7% of total p53312A/A cohort) developed liver tumors between 70 and 90 wk (Fig. 5A). In contrast, only 1 p53312A/+ mouse (1/20) (5.0% of remaining mice, 4.8% of total) developed liver hyperproliferation within this time frame. This was again found to be statistically significant (p53312A/A vs. p53312A/+, P = 0.0339; p53312A/A vs. p53WT, P = 0.0234) (Fig. 5A). The p53312A/A mice that developed liver tumors were mainly black (4/5 mice), and 4/5 of the liver tumors occurred in females, suggesting that there might be a gender and strain bias in this cohort for the development of liver tumors.
Fig. 5.
p53312A/A mice are more susceptible to liver tumors following exposure to IR. (A) Survival curve showing the incidence of liver tumors between 50 and 90 wk. (B) H&E staining of two tumors isolated from p53312A/A mice, showing the varied histology.
Histological analysis revealed that, although the hyperplasias in these mice were frequently benign, one liver displayed hepatocellular carcinoma (Fig. 5B, Lower Left and Lower Right) that had metastasized to the large intestine. The surrounding liver tissue in the afflicted mice was also frequently found to show circular, unstained areas consistent with the presence of lipid droplets. In addition, one p53312A/A mouse (female, black coat), killed at this point as it had developed a prolapse, was found to have a highly abnormal, fatty liver although it had not developed any hyperplasia. Therefore, a p53 Ser312Ala mutation can confer sensitivity to liver hyperplasia following IR.
p53 Ser312Ala Mutation Dampens the Transcriptional Activity of p53.
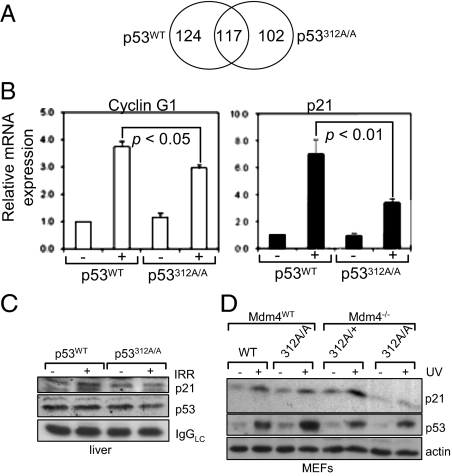
To better understand the mechanism by which Ser312 phosphorylation contributes to p53’s tumor suppression, we undertook a genome-wide gene array analysis using the Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay. Due to the increased incidence of liver tumors in p53312A/A mice, the analysis was carried out using mRNA from the livers of two pairs of p53WT and p53312A/A littermates that were either untreated or isolated 6 h after receiving a 4-Gy dose of IR. A p53 response has been previously detected in the liver at this time point (19) and is consistent with the phosphorylation of p53 at Ser312 seen in E1A-transformed MEFs (Fig. 1E). As expected, the expression levels of many genes, including some well-known p53 targets, were altered following IR (Table 2), although known p53-responsive genes such as Mdm2, Noxa, and Puma showed an insignificant change in expression after irradiation in both p53WT and p53312A/A mice. However, the spectrum of genes affected differed considerably between the two samples (Fig. 6A). There were 124 genes whose expression was changed in p53WT but not in p53312A/A mice; however, these genes were not recognized p53 target genes.
Table 2.
Known p53 target genes found to be differentially expressed in both p53wt and p53312A/A samples upon irradiation
| Gene symbol | Fold change (p53WT) | Fold change (p53312A/A) | Fold change (p53WT-p53312A/A) | |
| CDKN1A | 9.90 | 6.60 | −3.30 | p21 |
| CCNG1 | 3.25 | 2.15 | −1.10 | Cyclin G1 |
| GADD45G | 6.82 | 5.95 | −0.87 | Growth arrest and DNA damage-inducible 45 γ |
| PLK3 | 5.49 | 4.74 | −0.75 | Polo-like kinase 3 |
| PHLDA3 | 3.04 | 2.89 | −0.15 | Pleckstrin homology-like domain, family A, member 3 |
| BTG2 | 3.57 | 5.01 | 1.44 | B-cell translocation gene 2 |
These genes are among the 117 overlapping genes in Fig. 6A. Data indicate the fold change in each gene after irradiation.
Fig. 6.
p53 Ser312Ala mutation in the liver dampens the transcriptional activity of p53 and reduces p21 expression. Littermates were exposed to a 4-Gy IR dose, and the livers were isolated for analysis after 6 h. (A) Venn diagram showing the number of genes whose expression is altered in the liver after IR exposure. The expression of 117 genes was altered in both p53wt and p53312A/A mice, whereas 124 genes were altered only in p53WT mice and 102 genes were affected only in p53312A/A mice. (B) Real-time RT-PCR data (p21 and cyclin G1 transcripts) carried out in p53WT and p53312A/A liver RNA samples with and without IR. (C) Immunoblot showing reduced elevation in p21 protein expression in p53312A/A mice following IR. (D) p21 expression in p53WT and p53312A/A MEFs with or without Mdm4 following UV exposure (10 Jm−2).
The recognized p53-responsive genes whose expression differed to the greatest degree were p21 and cyclin G1 (Table 2). To validate our array data, quantitative RT-PCR was performed to measure the expression levels of p21 mRNA in p53WT versus p53312A/A mouse livers, with or without IR. Without IR, the levels of p21 and cyclin G1 mRNA in WT and mutant livers were comparable. However, after IR exposure, the increase in p21 and cyclin G1 mRNA was significantly less in p53312A/A mice than in WT mice (Fig. 6B and Table S2). In agreement, there was a small reduction in the level of p21 protein induced in p53312A/A livers in response to IR (Fig. 6C). This is further supported by the data obtained in p53wt versus p53312A/A MEFs (Fig. 6D), where p21 expression was lower with mutant p53, especially in the absence of Mdm4. These data suggest that phosphorylation at Ser312 enhances p53’s transcriptional activity.
By fine-tuning the actions of p53, Ser312 phosphorylation may have only a subtle effect on the expression of its target genes. However, when these effects are combined, they may be sufficient to mount a cellular response that is biologically significant following a genotoxic challenge such as IR. Our data demonstrate that despite possessing a mutation at only a single phosphorylation site, the Ser312Ala mutation confers increased susceptibility to tumorigenesis following IR exposure, especially in the thymus and, unusually, in the liver.
Discussion
In this study, we used knock-in mice carrying a Ser312Ala mutation in the p53 gene to investigate how phosphorylation at this site contributes to p53 function. As may be expected from a single amino acid change not known to be carcinogenic, these mice are viable and do not develop spontaneous tumors. This suggests that Ser312 does not influence p53’s role in development and life span under normal conditions and agrees with findings in similar mice: Ser18Ala (human Ser15), Ser23Ala (human Ser20), and Ser389Ala (human Ser392) knock-in mice all have minimal phenotypic abnormalities under normal conditions (20–22). The finding is also consistent with the hypothesis that phosphorylation of p53 may have a role in fine-tuning its regulation, evident only when p53 activity increases above a certain threshold.
However, following exposure to IR, p53 Ser312 mutant mice have a greater predisposition to tumors, particularly in the thymus and liver. The occurrence of thymic and T-cell lymphomas is common in mice with mutations in their p53 pathways, especially following irradiation (15, 18, 23). The time of onset for lymphomas in p53312A/A mice (26–52 wk) is comparable to that seen in p53+/− mice exposed to 4 Gy irradiation (20–50 wk) (15). However, unlike p53+/− mice, p53312A/A mice do not develop spontaneous lymphomas as tumors were seen only following irradiation.
Although we do not yet know the precise mechanism whereby Ser312Ala mutation causes thymic lymphomas, the fact that p53312A/A mice with a defined genetic background develop lymphomas relatively rapidly following IR demonstrates that Ser312 plays an important suppressive role in lymphoma development. This distinguishes p53312A/A mice from those with mutations at Ser18, Ser23, or Ser389, which do not develop lymphomas either spontaneously or following IR within a comparable time frame (21, 22, 24). p53312A/A mice are prone to thymic lymphomas after IR, yet their thymocytes are not resistant to apoptosis. Conversely, thymocytes derived from p53S23A and p53S18,23A mice are largely apoptosis-resistant, but the mice rarely develop thymic lymphomas (21, 22). Thus, lymphoma development is perhaps not always linked to apoptotic failure in DNA-damaged thymocytes. The other principal site for tumor development in p53312A/A mice is the liver. Liver tumors are unusual in p53-null and heterozygous mice following IR, although hepatocellular carcinomas were found in two mice with the p53R270H/− Li-Fraumeni mutation (25). This rarity may be due to the relatively rapid onset of other tumor types that mask the participation of the p53/p21 pathway in liver tumorigenesis (15, 18, 23, 26).
In our study, all of the p53312A/A mice that developed lymphomas had white coats, whereas the p53312A/A mice that acquired liver tumors were mostly black. This mixture of coat colors reflects their mixed genetic background and may explain the tissue distribution of the tumors seen. However, p53312A/A mice were more susceptible than either their WT or heterozygous counterparts of the same coat color to both lymphomas and liver tumors (Fig. S4). Importantly, C57BL/6J mice (black coats) are resistant to spontaneous liver tumor development (27), and FVB mice (white) have low lymphoma susceptibility (28). This strongly indicates that the mutation of p53, rather than the genetic background, sensitizes these mice to tumor development. The influence of strain on tumor outcome is well documented; e.g., p53-null mice develop teratocarcinomas in a 129/Sv background, whereas p53+/− BALB/c mice are susceptible to mammary tumors (29, 30). Subtle variations may exist between mouse strains in pathways modulated by irradiation that may influence tumor outcome. The precise nature of such differences is currently unknown and requires more investigation. Overall, these results reinforce the hypothesis that Ser312 positively contributes to tumor suppression by p53 and hint that the fine-tuning effect of a defined p53 phosphorylation site may be tissue and stimulus dependent.
As with the untransformed MEFs in Fig. 1E, we were unable to detect phosphorylation of p53 Ser312 in WT liver or thymus following IR, despite numerous attempts. However, Ser312 was phosphorylated in irradiated E1A-transformed MEFs, suggesting that phosphorylation at this site occurs in cells already undergoing chronic (in this case, oncogenic) stress. If exposure to IR causes such a stress in vivo, the failure of p53312A/A mice to phosphorylate Ser312 in these conditions may prevent p53 from efficiently counteracting this stress, making the mice susceptible to tumor growth.
Existing studies show that IR-induced p53 in liver does not induce apoptosis, but instead induces p21-mediated cell cycle arrest (19, 31, 32). Restoration of p53 in vivo also causes liver tumor regression via p53-mediated senescence (33). The fact that p53312A/A mice are prone to liver tumor development suggests that Ser312 mutation may hinder p53 by suppressing its ability to regulate cell cycle progression. This is supported by our observation that Ser312Ala mutation predisposes mice to lymphoma without affecting p53-mediated thymocyte apoptosis (Fig. 4 and Fig. S3). The reduced expression of p21 seen in the liver of p53312A/A mice, and MEFs derived from p53312A/A mice following DNA damage, correlates with previous reports that human p53 phosphorylated at Ser315 has an increased binding affinity for DNA (34). The resulting elevated expression of p21 is reversed by mutation of Ser315 to Ala or by inhibition of cdks by roscovitine (7, 11).
An increase in the expression of p21, but not proapoptotic genes such as Bax, has been reported in the livers of WT mice following IR exposure (32, 35). The elevated levels of p21 persisted for at least 10 wk, implying a central role for p21 in preventing proliferation following DNA damage in the liver (35). A reduction in p21 levels alongside altered p53 expression has also been observed in human hepatocellular carcinomas (36). Reduced p21 expression could also explain the partial rescue of the Mdm4 phenotype, as Mdm4−/−p21−/− embryos have a similar phenotype to that seen in the Mdm4−/−p53312A/+ mice (37). However, the number of genes whose expression varies between p53WT and p53312A/A mice (Fig. 6A) indicates that the difference in p21 expression is merely one element in a much more complex scenario. The fact that p21−/− mice do not develop liver tumors, either spontaneously or following IR exposure, would seem to confirm this (38).
This study has demonstrated that a single phosphorylation site is an important participant in tumor suppression by p53, depending upon which function is required and the site of suppression. Therefore, the many posttranslational modifications of p53 may act in a highly specific manner to subtly modify its behavior, and it is important that the contexts in which this occurs be determined.
Materials and Methods
Mice.
The p53312A/A knock-in mice were generated by inGenious Targeting Laboratory. Mdm4+/− mice were a generous gift from J.-C. Marine (University of Leuven) (12). All animal procedures were approved by local ethical review and licensed by the UK Home Office.
Antibodies.
The p53 phospho-Ser312 polyclonal antibody was made by immunizing rabbits with the peptide CTSApSPPQKKKP, followed by purification against the nonphosphorylated peptide to remove nonspecific antibodies. For more details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mark Shipman for his technical assistance and Dr. Claire Beveridge for critical reading of the manuscript. This work was funded by the Ludwig Institute for Cancer Research and the Association for International Cancer Research (X.L.), and the Italian Association for Cancer Research (G.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005165107/-/DCSupplemental.
References
- 1.Toledo F, Wahl GM. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 2.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 4.Radhakrishnan SK, Gartel AL. CDK9 phosphorylates p53 on serine residues 33, 315 and 392. Cell Cycle. 2006;5:519–521. doi: 10.4161/cc.5.5.2514. [DOI] [PubMed] [Google Scholar]
- 5.Katayama H, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff JR, Friedman PN, Marshak DR, Prives C, Beach D. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc Natl Acad Sci USA. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J. 2005;24:2768–2782. doi: 10.1038/sj.emboj.7600735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zacchi P, et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002;419:853–857. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- 9.Zheng H, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002;419:849–853. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- 10.Qu L, et al. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaydes JP, et al. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J Biol Chem. 2001;276:4699–4708. doi: 10.1074/jbc.M003485200. [DOI] [PubMed] [Google Scholar]
- 12.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 14.Finch RA, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- 15.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 16.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 17.Clarke AR, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 18.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 19.MacCallum DE, et al. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- 20.Bruins W, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol. 2004;24:8884–8894. doi: 10.1128/MCB.24.20.8884-8894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacPherson D, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689–3699. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–2622. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 24.Hoogervorst EM, et al. Lack of p53 Ser389 phosphorylation predisposes mice to develop 2-acetylaminofluorene-induced bladder tumors but not ionizing radiation-induced lymphomas. Cancer Res. 2005;65:3610–3616. doi: 10.1158/0008-5472.CAN-04-4328. [DOI] [PubMed] [Google Scholar]
- 25.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- 27.Stanley LA, et al. Proto-oncogene activation in liver tumors of hepatocarcinogenesis-resistant strains of mice. Carcinogenesis. 1992;13:2427–2433. doi: 10.1093/carcin/13.12.2427. [DOI] [PubMed] [Google Scholar]
- 28.Mahler JF, Stokes W, Mann PC, Takaoka M, Maronpot RR. Spontaneous lesions in aging FVB/N mice. Toxicol Pathol. 1996;24:710–716. doi: 10.1177/019262339602400606. [DOI] [PubMed] [Google Scholar]
- 29.Harvey M, McArthur MJ, Montgomery CA, Jr, Bradley A, Donehower LA. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J. 1993;7:938–943. doi: 10.1096/fasebj.7.10.8344491. [DOI] [PubMed] [Google Scholar]
- 30.Kuperwasser C, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–2159. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellamy CO, Clarke AR, Wyllie AH, Harrison DJ. p53 deficiency in liver reduces local control of survival and proliferation, but does not affect apoptosis after DNA damage. FASEB J. 1997;11:591–599. doi: 10.1096/fasebj.11.7.9212083. [DOI] [PubMed] [Google Scholar]
- 32.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 33.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pospísilová S, et al. Activation of the DNA-binding ability of latent p53 protein by protein kinase C is abolished by protein kinase CK2. Biochem J. 2004;378:939–947. doi: 10.1042/BJ20030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlik A, et al. Changes in transcriptome after in vivo exposure to ionising radiation reveal a highly specialised liver response. Int J Radiat Biol. 2009;85:656–671. doi: 10.1080/09553000903020024. [DOI] [PubMed] [Google Scholar]
- 36.Shi YZ, et al. Reduced p21(WAF1/CIP1) protein expression is predominantly related to altered p53 in hepatocellular carcinomas. Br J Cancer. 2000;83:50–55. doi: 10.1054/bjoc.2000.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinman HA, Sluss HK, Sands AT, Pihan G, Jones SN. Absence of p21 partially rescues Mdm4 loss and uncovers an antiproliferative effect of Mdm4 on cell growth. Oncogene. 2004;23:303–306. doi: 10.1038/sj.onc.1206925. [DOI] [PubMed] [Google Scholar]
- 38.Martín-Caballero J, Flores JM, García-Palencia P, Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–6238. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.