Abstract
Adaptive T-cell immunity relies on the recruitment of antigen-specific clonotypes, each defined by the expression of a distinct T-cell receptor (TCR), from an array of naïve T-cell precursors. Despite the enormous clonotypic diversity that resides within the naïve T-cell pool, interindividual sharing of TCR sequences has been observed within mobilized T-cell responses specific for certain peptide–major histocompatibility complex (pMHC) antigens. The mechanisms that underlie this phenomenon have not been fully elucidated, however. A mechanism of convergent recombination has been proposed to account for the occurrence of shared, or “public,” TCRs in specific memory T-cell populations. According to this model, TCR sharing between individuals is directly related to TCR production frequency; this, in turn, is determined on a probabilistic basis by the relative generation efficiency of particular nucleotide and amino acid sequences during the recombination process. Here, we tested the key predictions of convergent recombination in a comprehensive evaluation of the naïve CD8+ TCRβ repertoire in mice. Within defined segments of the naïve CD8+ T-cell repertoire, TCRβ sequences with convergent features were (i) present at higher copy numbers within individual mice and (ii) shared between individual mice. Thus, the naïve CD8+ T-cell repertoire is not flat, but comprises a hierarchy of recurrence rates for individual clonotypes that is determined by relative production frequencies. These findings provide a framework for understanding the early mobilization of public CD8+ T-cell clonotypes, which can exert profound biological effects during acute infectious processes.
The molecular mechanisms involved in T-cell receptor (TCR) generation permit an enormous array of combinatorial permutations with the potential to produce an estimated TCRαβ diversity exceeding 1015 in mice (1). In contrast, experimental estimates indicate that the actual murine naïve T-cell repertoire contains a mere 2 × 106 clonotypes, each with a population size of ∼10 cells (2). These numerical considerations suggest that the TCR repertoires of two genetically identical individuals should be largely dissimilar. However, various studies have demonstrated the presence of identical TCRβ chain sequences within antigen-experienced T-cell populations that target specific peptide–major histocompatibility complex (pMHC) epitopes in multiple individuals; this finding holds across species and extends to outbred macaque and human populations (3, 4). Irrespective of the selection processes that determine the recruitment of individual pMHC epitope-specific clonotypes into the memory T-cell pool, these observations indicate that such shared, or “public,” TCRs must first be present in the naïve T-cell repertoire of multiple individuals. Furthermore, it has been suggested that public TCR sequences might be generated more efficiently during the recombination process, thereby explaining differential interindividual frequencies of naïve T-cell clonotypes (5, 6).
Previously, we studied the immune TCRβ repertoires of CD8+ T-cell populations specific for virus-derived epitopes in mice (6), macaques (7), and humans (8). In all cases, the spectrum of TCRβ sharing across pMHC class I (pMHCI) epitope-specific CD8+ T-cell repertoires could be predicted on the basis of sequence production frequencies during VDJ recombination. In turn, TCRβ sequence production frequencies were dictated by the process of “convergent recombination” (4). According to this process, multiple recombination events converge to produce the same TCRβ nucleotide sequence, and multiple TCRβ nucleotide sequences converge to encode the same TCRβ amino acid sequence; in addition, multiple TCRβ amino acid sequences can converge to produce distinct TCRβ motifs.
The convergent recombination model leads to two fundamental predictions: (i) that multiple copies of frequently produced TCRs will be present in the naïve T-cell repertoire of any one individual, and (ii) that frequently produced TCRs will be present in the naïve T-cell repertoires of multiple individuals (4). In this study, we undertook a comprehensive quantitative analysis of the naïve CD8+ TCRβ repertoire in mice to test these predictions. The data confirm the key tenets of convergent recombination and show that this process is a major architect of the TCRβ repertoire within the naïve CD8+ T-cell pool.
Results
The Naïve TCR Repertoire Is Not Flat.
The naïve T-cell repertoire comprises a highly diverse population of clonotypes, each of which is present at a very low frequency. However, it is not clear whether all naïve T-cell clonotypes occur at similar frequencies, thereby producing a “flat” TCR landscape, or whether numerical differences exist such that a quantitative hierarchy of clonotypes is established. To investigate this issue, we conducted an intensive analysis of clonotype frequencies within narrow portions of the naïve murine TCR repertoire. We adopted this approach to ensure adequate sampling depth and minimize bias arising from lack of coverage, which would lead to an underestimation of frequency differences between individual clonotypes. The TRBV1/TRBJ2-1 and TRBV16/TRBJ2-5 gene rearrangements selected for this study are prevalent within memory CD8+ T-cell populations specific for the M38 SSPPMFRVP/H-2Kb (unpublished data) and IE-3 RALEYKNL/H-2Kb (9) epitopes derived from murine cytomegalovirus, respectively.
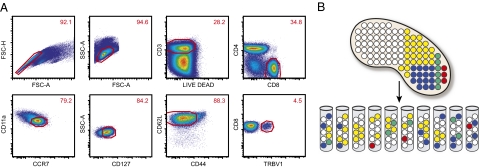
Naïve CD8+ T cells were stringently defined by polychromatic flow cytometry according to the expression of multiple lineage and phenotypic markers (Fig. 1A), thereby minimizing bias arising from inclusion of atypical memory CD8+ T cells with “naïve-like” phenotypes (10, 11) and the numerical advantage accrued by naïve CD8+ T-cell clonotypes that have undergone homeostatic proliferation (12, 13). Furthermore, each defined population was sorted at >98% purity by flow cytometry into multiple aliquots, each of which was processed independently to generate quantal TCR sequence data, as described in Methods (Fig. 1B). This experimental design was used to circumvent the molecular bias that favors high-frequency templates during the exponential phase of amplification, even in PCRs that incorporate only single primer pairs, and to maximize the detection sensitivity for low-frequency clonotypes. Thus, the number of aliquots in which a given TCRβ sequence was detected provides an accurate measure of relative frequency for that clonotype independent of quantitative measurements within individual PCRs. In addition, this approach eliminates bias introduced by differences in the number of TRB gene transcripts per cell. At least 70 TCRβ sequences per aliquot were obtained from a minimum of 22 aliquots for each mouse, yielding a total of >1,900 TCRβ sequences from each of three mice (Table 1).
Fig. 1.
Naïve CD8+ T-cell repertoire sampling strategy. (A) Representative gating scheme for flow cytometric sorting of TRBV1+ naïve CD8+ T cells from mouse spleen. (Upper) Bivariate plots delineating the sequential selection of singlet, live CD3+CD4−CD8+ lymphocytes. (Lower) Bivariate plots showing the phenotypic markers used to define naïve CD8+ T cells within this population before sorting based on TCRVβ1 expression. A corresponding approach was adopted for the isolation of splenic TRBV16+ naïve CD8+ T cells from mouse 3. (B) Schematic representation of sample partitioning. In this cartoon, a mouse spleen contains 100 TCRVβ-defined naïve CD8+ T-cell clonotypes. White dots represent single copies of unique clonotypes; colored dots represent multiple copies of the same clonotype. The TCRVβ-defined naïve CD8+ T cells are sorted randomly into 10 collection tubes; accordingly, the number of aliquots in which any given clonotype is detected provides a measure of its original clonal frequency in the spleen.
Table 1.
Characteristics of naïve TCRβ repertoire samples
| Mouse 1 TRBV1/TRBJ2-1 | Mouse 2 TRBV1/TRBJ2-1 | Mouse 3 TRBV16/TRBJ2-5 | |
| Total no. of TCR sequences | 1,952 | 1,911 | 2,098 |
| No. of unique CDR3 a.a. sequences | 1,193 | 1,178 | 1,233 |
| No. of unique CDR3 n.t. sequences | 1,356 | 1,338 | 1,402 |
| No. of aliquots | 22 | 22 | 25 |
| No. of TCRs per aliquot, mean | 88.7 | 86.9 | 83.9 |
| No. of TCRs per aliquot, range | 79–94 | 78–94 | 70–92 |
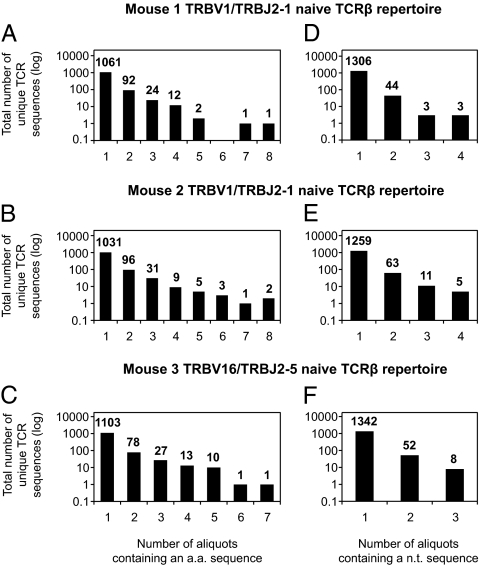
For each mouse, the clonotypic precursor frequency of the observed TCRβ amino acid and nucleotide sequences was estimated based on the number of aliquots in which the TCRβ sequence appeared (Fig. 2). Importantly, this defines the minimum number of times that each TCRβ sequence was produced. For the naïve TRBV1/TRBJ2-1 repertoires in mouse 1 and mouse 2, the number of aliquots in which individual TCRβ amino acid and nucleotide sequences were detected spanned ranges up to a maximum of eight (Fig. 2 A and B) and four (Fig. 2 D and E), respectively. Similarly, for the naïve TRBV16/TRBJ2-5 repertoire in mouse 3, individual TCRβ amino acid sequences were found in up to seven aliquots, and individual TCRβ nucleotide sequences were found in up to three aliquots (Fig. 2 C and F). Thus, independent of the selected TRB gene rearrangements, a clear hierarchy of clonotypic precursor frequencies for different TCRβ amino acid and nucleotide sequences was present in each individual mouse for each portion of the sampled naïve repertoire.
Fig. 2.
Prevalence of unique TCRβ clonotypes in sampled naïve CD8+ T-cell repertoires. The distributions of unique TCRβ amino acid (a.a.) sequences (A–C) and nucleotide (n.t.) sequences (D–F) are shown across samples of the depicted naïve CD8+ T-cell repertoires for mouse 1 (A and D), mouse 2 (B and E), and mouse 3 (C and F).
TCRβ Sequences with High Clonotypic Precursor Frequencies Incorporate Fewer Nucleotide Additions.
Previous analyses of interindividual TCR sharing in antigen-experienced epitope-specific CD8+ T-cell responses to infection have clearly demonstrated that the extent of TCR sharing between individuals is related to TCR sequence production frequency, which in turn is determined on a probabilistic basis during gene recombination (4, 6–8). If this process of convergent recombination were a determinant of antigen-driven clonotype representation within the memory CD8+ T-cell pool, then a unified theory would predict that such findings hold within the naïve CD8+ T-cell pool across individuals irrespective of specificity.
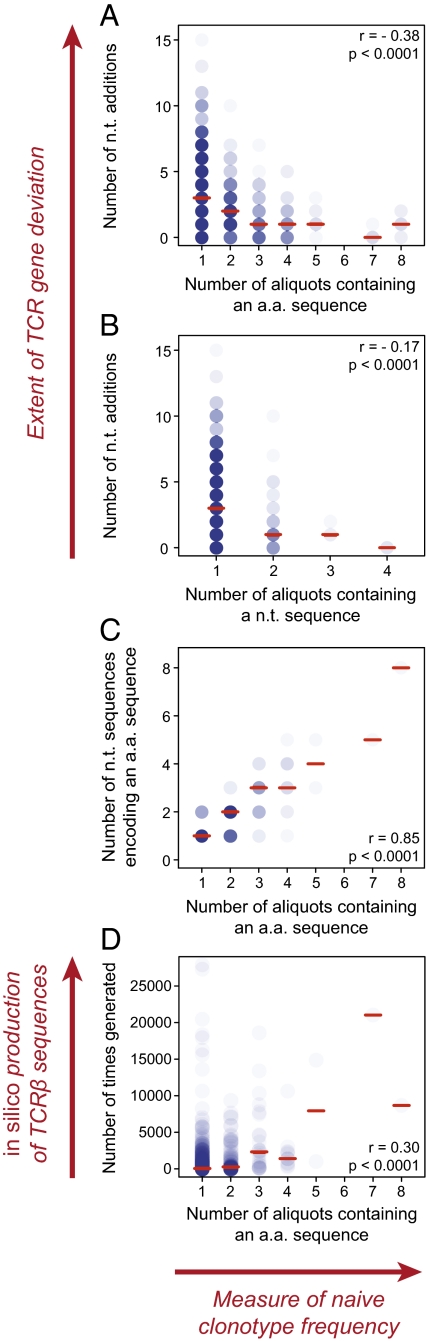
One major indicator of TCR sequence production efficiency is the number of nucleotide additions required to generate that particular sequence. Thus, TCR sequences with no or few nucleotide additions should be easily made and commonly generated; conversely, TCR sequences should become progressively more difficult to make and relatively less frequent as the required number of nucleotide additions increases. Consistent with these predictions, TCRβ amino acid and nucleotide sequences that were present in greater numbers of aliquots after partitioning of the sampled naïve CD8+ T-cell repertoires could be made with fewer nucleotide additions (Fig. 3 A and B and Fig. S1 A–F). The most frequently observed TCRβ amino acid sequences in the naïve repertoires of all three mice could be made with no more than two nucleotide additions; in contrast, >10 nucleotide additions were required to make some of the sequences that were detected in only one aliquot. Moreover, the negative correlation between the number of aliquots in which a TCRβ amino acid or nucleotide sequence was present and the minimum number of nucleotide additions was significant in all three mice (Fig. 3 A and B and Fig. S1 A–F; P < 0.0001, Spearman's rank correlation test).
Fig. 3.
Features of the observed TRBV1/TRBJ2-1 naïve CD8+ T-cell repertoire in mouse 1. (A and B) Relationship between the minimum number of nucleotide additions and the number of aliquots in which a TCRβ amino acid (a.a.) sequence (A) or nucleotide (n.t.) sequence (B) was detected. (C) Relationship between the number of nucleotide sequences observed to encode each unique amino acid sequence and the number of aliquots in which a TCRβ amino acid sequence was detected. (D) The production of 2 × 107 in-frame TCRβ sequences was simulated; the number of times that each amino acid sequence was generated in silico is plotted against the number of aliquots in which the same sequence was observed in vivo. Each circle represents an observed TCRβ sequence. The opacity of the circles indicates the density of the points plotted, and the red lines indicate the median values. Correlations are based on Spearman's rank correlation test.
TCRβ Amino Acid Sequences with High Clonotypic Precursor Frequencies Are Produced by Multiple Nucleotide Sequences.
Codon degeneracy enables multiple nucleotide sequences to encode for the same amino acid sequence. Accordingly, a second major indicator of TCR sequence production efficiency is the number of nucleotide sequences that can encode for that particular amino acid sequence. In all three mice, TCRβ amino acid sequences that were present in greater numbers of aliquots were encoded by proportionately greater numbers of different nucleotide sequences (Fig. 3C and Fig. S1 G–I). The most frequently observed TRBV1/TRBJ2-1 amino acid sequence in mouse 1, CTCSAGTGGYAEQFF, was detected in eight aliquots and was encoded by eight different nucleotide sequences. In mouse 2, the same TCRβ amino acid sequence was detected in six aliquots, encoded by five different nucleotide sequences. Similarly, the most frequent TRBV1/TRBJ2-1 amino acid sequences in mouse 2, CTCSAGGNYAEQFF and CTCSAGQYAEQFF, were detected in eight aliquots and were encoded by six and five different nucleotide sequences, respectively. In mouse 1, both of these TCRβ amino acid sequences were detected in four aliquots. Interestingly, in mouse 2, the TRBV1/TRBJ2-1 amino acid sequence CTCSAGGYAEQFF was detected in seven aliquots but was encoded by only two nucleotide sequences, neither of which required additions. In mouse 3, the most frequent TRBV16/TRBJ2-5 amino acid sequence, CASSLVGGQDTQYF, was detected in seven aliquots and was encoded by four different nucleotide sequences. In all three mice, significant positive correlations were found between the number of aliquots in which a TCRβ amino acid sequence was present and the number of encoding nucleotide sequences (Fig. 3C and Fig. S1 G–I; P < 0.0001, Spearman's rank correlation test).
TCRβ Sequences with High Clonotypic Precursor Frequencies Are Generated More Easily by Convergent Recombination.
The results reported in the previous sections suggest that TCRβ sequences present at higher frequencies in the naïve repertoire have the potential to be made more efficiently by VDJ recombination; specifically, they tend to be encoded by a greater variety of nucleotide sequences, and these sequences tend to require fewer nucleotide additions (Fig. 4). These features indicate that convergent recombination plays an important role in the efficiency of TCRβ sequence production, and that this is a major determinant of clonotype frequency in the naïve TCRβ repertoire. It is not possible to determine the actual recombination events that produced any observed TCRβ sequence, however. Thus, a frequently detected TCRβ sequence might have been produced many times by the same recombination mechanism or several times by each of multiple different recombination mechanisms. To determine if the higher-frequency TCRβ sequences that we observed in the sampled naïve CD8+ T-cell repertoires could be generated more efficiently by unbiased gene recombination, we used computer simulations of a random VDJ recombination process.
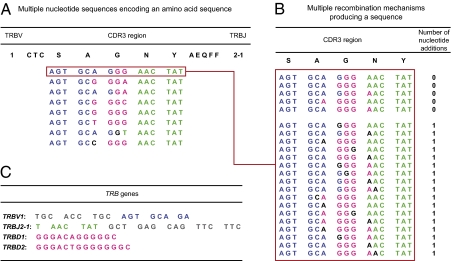
Fig. 4.
Representation of convergent recombination in the TRBV1/TRBJ2-1 naïve CD8+ T-cell repertoire. Convergent recombination is illustrated for the amino acid sequence CTCSAGNYAEQFF, which was detected in seven aliquots in mouse 1 and in six aliquots in mouse 2. (A) This TCRβ clonotype was encoded by a total of eight different nucleotide sequences across both repertoires. One possible alignment with TRBV1 (blue), TRBD1/TRBD2 (pink), and TRBJ2-1 (green) genes involving the minimal number of nucleotide additions (black) is shown. (B) At a second level of convergent recombination, multiple recombination mechanisms with similarly low numbers of nucleotide additions can produce the same nucleotide sequence. This is shown for one of the nucleotide sequences (red box) encoding CTCSAGNYAEQFF. (C) The mouse TRB gene sequences used.
The generation of 2 × 107 in-frame sequences was simulated using the TRBV1/TRBJ2-1 and TRBV16/TRBJ2-5 gene combinations. Of the potential unique TCRβ sequences observed per mouse that could be produced within the simulation parameters, means of 92.0% and 64.1% were generated in silico at the amino acid and nucleotide levels, respectively. Analysis of these simulated TCRβ repertoires revealed significant positive correlations between the number of aliquots per mouse in which each TCRβ amino acid or nucleotide sequence was observed and the number of times that the TCRβ sequences were generated in silico (Fig. 3D and Fig. S2 A–F; P < 0.0001, Spearman's rank correlation test). The role of convergent recombination in TCRβ clonotype production efficiency was further supported by significant positive correlations (P < 0.0001, Spearman's rank correlation test) between (i) the number of different nucleotide sequences encoding an amino acid sequence in silico and the number of aliquots per mouse in which the TCRβ amino acid sequence was detected (Fig. S2 G–I) and (ii) the number of different VDJ recombination mechanisms (i.e., different splicings of the TCRβ genes and nucleotide additions) producing each amino acid and nucleotide sequence in silico and the number of aliquots per mouse in which the TCRβ amino acid or nucleotide sequence was detected (Fig. S2 J–O). Thus, variable TCRβ sequence production frequencies occurred even in an unbiased process of TRBV, TRBD, and TRBJ gene recombination. Furthermore, in silico TCRβ production frequencies were significantly correlated with observed TCRβ sequence frequencies within the naïve CD8+ T-cell repertoire.
TCRβ Sequences with High Clonotypic Precursor Frequencies Are Shared Between Mice.
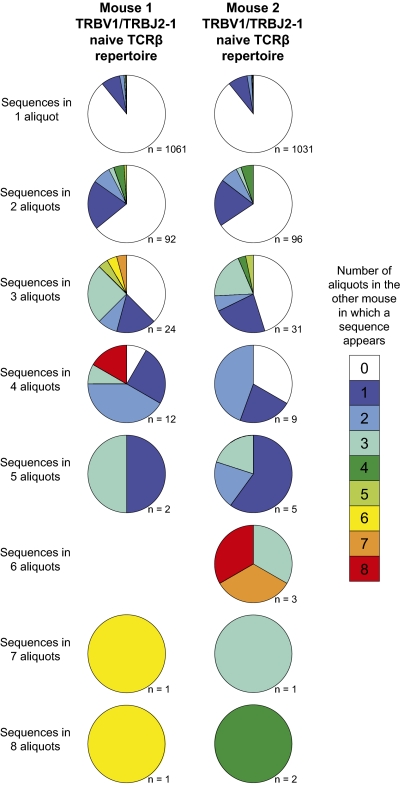
If convergent recombination determines, at least in part, interindividual TCR sharing within antigen-specific memory CD8+ T-cell responses, then sequence production efficiency must play a comparable role in shaping observed clonotype frequencies within the naïve CD8+ T-cell repertoires across individuals. Sharing of naïve TRBV1/TRBJ2-1 clonotypes was observed between mouse 1 and mouse 2. Of 2,193 different TCRβ amino acid sequences found across both mice, 178 (8.1%) were shared between mice (Fig. 5 and Fig. S3). Similarly, of the 2,622 different TCRβ nucleotide sequences found across both mice, 72 (2.7%) were shared between mice. Importantly, the sharing of TCRβ sequences between mice was strongly predicted by the frequency of these sequences within individual mice. Thus, a greater proportion of the TCRβ amino acid and nucleotide sequences that were detected in numerous aliquots per mouse were observed in the other mouse as well (Fig. S3 A–D). For example, only 10.8% of the 1,061 TCRβ amino acid sequences that were detected in one aliquot in mouse 1 were present in mouse 2. In contrast, 100% of the TCRβ amino acid sequences that were present in five or more aliquots in either mouse were also found in the other mouse. Furthermore, there was a significant correlation between mouse 1 and mouse 2 in terms of the number of aliquots in which each shared TCRβ amino acid sequence was detected (Fig. S3E; P < 0.0001, Spearman's rank correlation test).
Fig. 5.
The relationship between TRBV1/TRBJ2-1 naïve CD8+ T-cell clonotype frequency and sharing between mice. Each pie chart represents all TCRβ amino acid sequences detected in a given number of aliquots in one mouse. The size of each pie slice indicates the proportion of those TCRβ sequences present in the other mouse; colors indicate the number of aliquots in which each sequence was detected in the other mouse, as depicted in the key.
To determine a direct association between the sharing of naïve TCRβ sequences and their potential production efficiency during VDJ recombination, we compared the features of TCRβ sequences found in one or both mice. TCRβ amino acid and nucleotide sequences found in both mouse 1 and mouse 2 required significantly fewer nucleotide additions compared with those TCRβ sequences found in only one mouse (Fig. S4 A and B; P < 0.0001, Mann–Whitney U test). In addition, TCRβ amino acid sequences found in both mice were encoded by significantly higher numbers of different nucleotide sequences compared with those TCRβ sequences found in only one mouse (Fig. S4C; P < 0.0001, Mann–Whitney U test).
Discussion
In this study, we analyzed segments of the naïve CD8+ T-cell repertoire in mice defined by the TRBV1/TRBJ2-1 and TRBV16/TRBJ2-5 gene combinations to test the fundamental predictions of convergent recombination. Consistent with this model, both naïve TCRβ frequencies and interindividual TCRβ sharing within the naïve CD8+ T-cell pool were found to be directly related to predicted TCRβ sequence production frequencies. Thus, TCRβ sequences with higher intraindividual clonotypic precursor frequencies that were also shared between mice contained fewer additions at the nucleotide level and were encoded by multiple alternative nucleotide sequences at the amino acid level. Furthermore, these TCRβ sequences were produced more readily in computer simulations of a random VDJ recombination process and the relative frequencies generated in silico correlated strongly with those observed in vivo. These data establish convergent recombination as a major determinant of the naïve CD8+ T-cell landscape.
Key elements of the approach adopted here included the rigorous definition of naïve CD8+ T cells, high purity flow cytometric sorting and the partitioning of isolated populations for independent and parallel molecular processing. Combined with in-depth sequence analysis, these features enabled the accurate measurement of individual CD8+ T-cell frequencies within defined segments of the naïve repertoire. However, the naïve CD8+ T-cell repertoire is shaped not only by genetic recombination mechanisms, but also by thymic selection and possible peripheral expansion. The latter possibility was limited by the inclusion of specific cell surface markers during flow cytometric analysis and sorting to minimize the influence of homeostatic proliferation (Fig. 1), although it remains feasible that differential clonotypic division within the sampled populations could introduce a source of bias. In addition, it is likely that not all frequently produced TCRβ sequences survive thymic selection to enter the peripheral naïve repertoire. Indeed, certain TCRβ sequences that were frequently generated in silico were either not detected or present only rarely and not shared; this could reflect the production frequency and pairing promiscuity of TCRα chain partners, negative selection in the thymus, or homeostatic proliferation associated with differentiation. Nonetheless, despite these considerations, the data reported herein clearly show that clonotypic precursor frequency and sharing can be predicted by production frequency. Thus, the underlying mechanism of convergent recombination remains a primary determinant of TCRβ architecture within the naïve CD8+ T-cell pool irrespective of other influences, including the potential for differential TCRα pairing. This observation is consistent with a recent study, which found no evidence that public clonotypes were preferentially selected in the thymus (14). In contrast, another study reported a “raw” TCR repertoire generated by random gene rearrangements that was flat with few repeated sequences, with hierarchical shape attributed instead to thymic selection (15). Given the estimated size of the peripheral naïve repertoire, however, the sequencing depth in this latter study might have been insufficient to observe the true extent of clonotypic frequency differences within the preselection repertoire.
We previously demonstrated that convergent recombination plays an important role in TCRβ sharing between individuals within antigen-experienced epitope-specific CD8+ T-cell responses (4, 6–8). Coupled with the current data, these studies demonstrate that the hierarchy of TCRβ clonotype frequencies established by VDJ recombination is maintained through thymic selection into the naïve repertoire and plays an important role in shaping the clonotypic composition of antigen-specific memory CD8+ T-cell populations. Consistent with these findings, public clonotypes specific for an immunodominant influenza virus-derived epitope have been shown to exist at relatively high frequencies in the preimmune cognate TCRβ repertoire of uninfected mice (16). Thus, a coherent explanation emerges for the enigmatic phenomenon of TCR “publicity” within adaptive T-cell immune responses. During the initial phases of antigen challenge, different pMHCI complexes will recruit different cognate clonotypes into the specific memory and effector CD8+ T-cell pools. This priming process is highly efficient (17), even for clonotypes that exhibit low levels of functional reactivity with the incoming antigen (18). Accordingly, all clonotypes that exceed a minimal antigen avidity threshold will be mobilized from the naïve CD8+ T-cell repertoire. Consequently, initial recruitment will depend primarily on the presence and availability of clonotypes that meet this minimal requirement within the naïve CD8+ T-cell repertoire. Convergent clonotypes, which are present at high frequencies in the naïve repertoires of multiple individuals due to the pseudorandom mechanisms of TCR gene rearrangement, will thus have a kinetic advantage and dominate early CD8+ T-cell responses in any given individual with the appropriate MHCI restriction element. Notably, this process will operate regardless of subsequent pressures that might modulate clonotype frequency hierarchies within pMHCI epitope-specific CD8+ T-cell populations, such as ongoing avidity-based selection in the presence of persistent pathogen-derived antigens (19).
Public clonotypes appear to play important roles during the early phases of infection with intracellular pathogens and can mediate either adverse or protective effects. This dichotomy is epitomized by studies of SIV infection in Mamu-A*01+ rhesus macaques. The earliest CD8+ T-cell response, which is specific for the Tat TL8/Mamu-A*01 epitope, is highly public and comprises TCRβ sequences that converge to produce a CDR3β motif. These features facilitate viral immune escape by epitope mutation at presumed TCR contact residues (20). In contrast, a more clonotypically diverse and protective CD8+ T-cell response specific for the biologically constrained Gag CM9/Mamu-A*01 epitope emerges marginally later. The degree of protection conferred by this response is predicted by the number of initially recruited public clonotypes, which presumably are mobilized more rapidly due to higher precursor frequencies within the naïve repertoire to control viral replication and mitigate the ensuing pathological consequences (21). Our data provide a molecular basis for the generation and selection of these biologically important CD8+ T-cell clonotypes, with attendant implications for T cell-based immune interventions.
Methods
Animals.
Female C57BL/6 mice (Jackson Laboratory) were maintained in specific pathogen-free conditions at the Vaccine Research Center Animal Care Unit, National Institute of Allergy and Infectious Diseases, National Institutes of Health. All experiments were approved by the Vaccine Research Center's Animal Care and Use Committee and were conducted in accordance with the policies and regulations of the National Institutes of Health. Mice were 24 wk old at the time of organ harvest.
Isolation of Naïve Splenic CD8+ T Cells with Specific TCRVβ Usage.
Entire spleens were harvested, crushed, and washed through 40-μm polystyrene cell filters with PBS. After two more washes in PBS and lysis of red blood cells using ACK buffer (Gibco), splenocytes were stained with the following monoclonal antibodies (mAbs) according to standard procedures: (i) αCD3-PacificBlue, αCD4-Alexa700, αCD8-APC-H7, αCD11a-FITC, αCD44-Cy5PE, and αCD62L-Cy7PE (BD Biosciences); (ii) αCCR7-biotin and αCD127-Alexa647 (eBiosciences); and (iii) either αTCRVβ2-PE (BD Biosciences) or αTCRVβ11-PE (BioLegend). Streptavidin-QD655 (Invitrogen) was used to visualize CCR7 staining. Dead cells were excluded using LiveDead Aqua (Molecular Probes). Naïve CD8+ T cells expressing either TCRVβ1 or TCRVβ16, which correspond to TCRVβ2S1 and TCRVβ11S1, respectively, in Arden's nomenclature (22), were sorted to >98% purity using a modified FACSAria flow cytometer (BD Immunocytometry Systems). Aliquots of 5,000 sorted cells were collected directly into 1.5-mL microfuge tubes (Sarstedt) containing 100 μL of RNAlater (Applied Biosystems).
RNA Extraction and cDNA Synthesis.
Total mRNA was extracted from each aliquot of cells using the Oligotex Direct mRNA Mini Kit (Qiagen). First-strand cDNA synthesis was performed using a modified version of the SMART procedure (switching mechanism at the 5′ end of RNA transcript) with RACE (rapid amplification of cDNA ends) as described previously (23).
Amplification of Specific TRBV/TRBJ Gene Rearrangements.
Primers were designed de novo or derived from previous reports (24). The following sequences were used: 5′-GGGGACAAAGAGGTCAAATC-3′ (mTRBV1), 5′-ACCAGGGACACGACTCACCGTCC-3′ (mTRBJ2.1), 5′-GCTCAGATGCCCAATCAGTCGCA-3′ (mTRBV16) and 5′-CCAGGCACTCGGCTCCTCGT-3′ (mTRBJ2.5). For amplification of specific TRBV/TRBJ gene rearrangements, each PCR contained 1× HiFi Buffer, 3 mM MgSO4, 200 μM dNTPs, 2.8 U platinum Taq HiFi DNA polymerase (Invitrogen), and 10 pmol of each primer in a final volume of 50 μL. Thermocycling conditions were 95 °C for 30 s, 58 °C for 30 s, and 68 °C for 30 s. Each PCR was conducted over 30 cycles following a single step at 95 °C for 5 min, and elongation was completed at 72 °C for 60 s. Amplicons were purified, cloned, and sequenced using standard Sanger-based technology as described previously (19).
TCR Gene Alignment and Estimation of Nucleotide Additions.
Analysis of TCRβ sequences was conducted with reference to the *01 allele sequences for murine TRBV, TRBD, and TRBJ genes, which were obtained from the international ImMunoGeneTics (IMGT) information system (http://imgt.cines.fr/); the IMGT nomenclature is used throughout for all TCR genes (25). Expressed TCRβ sequences were aligned with germline TRB genes using the pairwise alignment that produced the greatest percentage match. The TRBV and TRBJ genes were sequentially aligned to the 5′ and 3′ ends of the TCRβ sequence, respectively. Unaligned sequence in the TRBV/TRBJ junctional region was attributed to a TRBD gene for the longest consecutive match of at least two nucleotides. Any further unaligned nucleotides at the TRBV/TRBD and TRBD/TRBJ junctions were considered nucleotide additions.
Identity of TCRβ Sequences.
A TCRβ sequence was considered to be present in two aliquots or shared between two individual mice if an identical CDR3β sequence (including TRBV and TRBJ gene usage) was found in the repertoires of both aliquots/mice.
Simulation of VDJ Recombination.
The relative production frequencies of observed TCRβ sequences were estimated using computer simulations of a random VDJ recombination process (6). Simulation parameters were chosen to allow for production of the majority of observed TCRβ sequences. The simulated VDJ recombination process involved a randomly determined number, from 0 to 12, of nucleotide deletions from both the 3′ end of the TRBV gene and the 5′ end of the TRBJ gene. Involvement of either the TRBD1 or TRBD2 gene was determined randomly; nucleotide deletions from both the 5′ and 3′ ends of the TRBD gene were allowed up to the full length of the gene. A randomly determined number of nucleotides, again from 0 to 12, were then added at the TRBV/TRBD and TRBD/TRBJ gene segment junctions. There were 10, 7, and 8 TCRβ clonotypes from the observed repertoires of mouse 1, mouse 2, and mouse 3, respectively, that were omitted from analysis of the in silico TCRβ repertoires; these TCRβ clonotypes required greater than 12 nucleotide additions or deletions and thus could not be produced in the simulations. Of the 2 × 107 in-frame sequences generated in silico, a total of 5.8% (mouse 1 and mouse 2) and 2.6% (mouse 3) of the simulated TRBV1/TRBJ2-1 and TRBV16/TRBJ2-5 amino acid sequences, respectively, were observed in vivo. Computer simulations were performed using Matlab 7.9.0 (Mathworks).
Statistical Analysis.
Statistical analyses were conducted using GraphPad Prism (GraphPad Software). Correlations were performed using Spearman's rank correlation test. Comparisons between two groups were performed using the Mann–Whitney U test.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council (MRC) UK, the Australian Research Council (ARC), the Australian National Health and Medical Research Council (NHMRC), and the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health. D.A.P. is an MRC Senior Clinical Fellow, M.F.Q. is a Marie Curie International Outgoing Fellow, M.P.D. is an NHMRC Senior Research Fellow, and V.V. is an ARC Future Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010586107/-/DCSupplemental.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Casrouge A, et al. Size estimate of the alpha beta TCR repertoire of naïve mouse splenocytes. J Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 3.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 4.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 5.Fazilleau N, et al. Valpha and Vbeta public repertoires are highly conserved in terminal deoxynucleotidyl transferase–deficient mice. J Immunol. 2005;174:345–355. doi: 10.4049/jimmunol.174.1.345. [DOI] [PubMed] [Google Scholar]
- 6.Venturi V, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci USA. 2006;103:18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venturi V, Chin HY, Price DA, Douek DC, Davenport MP. The role of production frequency in the sharing of simian immunodeficiency virus–specific CD8+ TCRs between macaques. J Immunol. 2008;181:2597–2609. doi: 10.4049/jimmunol.181.4.2597. [DOI] [PubMed] [Google Scholar]
- 8.Venturi V, et al. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J Immunol. 2008;181:7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- 9.Jones M, et al. IL-10 restricts memory T cell inflation during cytomegalovirus infection. J Immunol. 2010;185:3583–3592. doi: 10.4049/jimmunol.1001535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Precopio ML, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haluszczak C, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada K, Jameson SC. Naïve T cell homeostasis: From awareness of space to a sense of place. Nat Rev Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 14.Furmanski AL, et al. Public T cell receptor beta-chains are not advantaged during positive selection. J Immunol. 2008;180:1029–1039. doi: 10.4049/jimmunol.180.2.1029. [DOI] [PubMed] [Google Scholar]
- 15.Correia-Neves M, Waltzinger C, Mathis D, Benoist C. The shaping of the T cell repertoire. Immunity. 2001;14:21–32. doi: 10.1016/s1074-7613(01)00086-3. [DOI] [PubMed] [Google Scholar]
- 16.La Gruta NL, et al. Primary CTL response magnitude in mice is determined by the extent of naïve T cell recruitment and subsequent clonal expansion. J Clin Invest. 2010;120:1885–1894. doi: 10.1172/JCI41538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Heijst JW, et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325:1265–1269. doi: 10.1126/science.1175455. [DOI] [PubMed] [Google Scholar]
- 18.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price DA, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price DA, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Price DA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 23.Douek DC, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 24.Kato T, et al. Comparison of the J beta gene usage among different T cell receptor V beta families in spleens of C57BL/6 mice. Eur J Immunol. 1994;24:2410–2414. doi: 10.1002/eji.1830241022. [DOI] [PubMed] [Google Scholar]
- 25.Lefranc MP, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.