Abstract
The molecular mechanisms of translation termination and mRNA surveillance in archaea remain unclear. In eukaryotes, eRF3 and HBS1, which are homologous to the tRNA carrier GTPase EF1α, respectively bind eRF1 and Pelota to decipher stop codons or to facilitate mRNA surveillance. However, genome-wide searches of archaea have failed to detect any orthologs to both GTPases. Here, we report the crystal structure of aRF1 from an archaeon, Aeropyrum pernix, and present strong evidence that the authentic archaeal EF1α acts as a carrier GTPase for aRF1 and for aPelota. The binding interface residues between aRF1 and aEF1α predicted from aRF1·aEF1α·GTP ternary structure model were confirmed by in vivo functional assays. The aRF1/eRF1 structural domain with GGQ motif, which corresponds to the CCA arm of tRNA, contacts with all three structural domains of aEF1α showing striking tRNA mimicry of aRF1/eRF1 and its GTPase-mediated catalysis for stop codon decoding. The multiple binding capacity of archaeal EF1α explains the absence of GTPase orthologs for eRF3 and HBS1 in archaea species and suggests that universal molecular mechanisms underlie translational elongation and termination, and mRNA surveillance pathways.
Keywords: genetic code, protein quality control, tRNA mimicy
In bacteria, eukaryotes and archaea, the stop codons are recognized by the tRNA mimicking class-I release factors (RFs) (1). The bacterial class-I RFs, RF1 and RF2, recognize the UAG/UAA and UGA/UAA stop codons, respectively, by their conserved tripeptide recognition motifs (2) and catalyze polypeptide release by their conserved GGQ motifs (3). The crystal structures of the Thermus thermophilus 70S ribosome complexed with RF1 or RF2 (4, 5) demonstrated that the functional domains of bacterial RFs work at the ribosomal catalytic sites corresponding to the tRNAs by a mechanism of “molecular mimicry.” By contrast, in archaea and eukaryotes, a single class-I release factor, aRF1 and eRF1, respectively, recognizes all three stop codons and catalyzes polypeptide-chain release (6, 7). Although eRF1 and aRF1 share highly conserved sequence motifs throughout the molecule (8) (Fig. S1), they lack phylogenetic and structural similarities with the bacterial class-I release factors, except for the universally conserved GGQ motifs (3) and their overall tRNA-like configurations (9).
eRF1 forms a heterodimeric complex with class-II release factor, eRF3, an elongation factor 1 alpha (EF1α)-related essential factor (Fig. S2), to complete the overall translation termination process in a GTP-dependent manner (10, 11). Recently, the crystal structures of human and Schizosaccharomyces pombe full-length eRF1 in complex with eRF3 lacking the GTP binding domain were solved, revealing important molecular details, including one of the binding sites between eRF1 and eRF3 (hereafter called “site 1”) (10). The binding of eRF1 and eRF3 at site 1 occurs primarily without GTP, and the small-angle X-ray scattering (SAXS) analysis of the eRF1·eRF3·GTP complex predicted a eRF1·eRF3·GTP complex model (10), in which regulatory contact(s) are formed between another structural domain of eRF1 and the GTP regulatory regions of eRF3, which are situated outside of site 1. Although the precise contact sites between eRF1 and eRF3 are not clear, the overall shape of the eRF1·eRF3·GTP complex resembles that of the tRNA·EF-Tu·GTP complex structure, strongly suggesting the existence of structural and functional analogies between the elongation and termination steps in eukaryotes (10). However, the structural details for the GTPase-mediated decoding of stop codons by eRFs and the similarities to the tRNA system remain to be clarified.
Recently, the atomic structures of the eRF1/aRF1-related Pelota proteins (Dom34p in budding yeast) were reported (12, 13). Pelota binds Hbs1, which is another EF1α-related subfamily GTPase (Fig. S2), and is considered to play a crucial role in mRNA surveillance for protein synthesis (14). In addition to the conservation of eRF1/aRF1 and Pelota/aPelota in eukaryotes and archaea, the phylogenetic absence of eRF3 and Hbs1 orthologs in archaea revealed by systematic genome analyses (8), raised intriguing questions about the identities of the molecular basis for in translation termination and mRNA surveillance between eukaryotes and archaea.
Here we present the atomic structure of aRF1 from a Crenarchaeon, Aeropyrum pernix (ape-aRF1). The fact that the ape-aRF1 maintained the putative GTPase binding domain structure (Fig. 1), which was identified in eukaryotes, prompted us to test the hypothesis that the authentic archaeal EF1α (aEF1α) could serve as a carrier GTPase protein for both aRF1 and aPelota onto the ribosome. Our biochemical and genetic analyses provided the evidence of the functional binding between aRF1/aPelota and aEF1α, which explains the absence of specific GTPases for the two tRNA mimicking proteins in archaeal species. Moreover, based on our crystal structure of the aPelota·aEF1α·GTP complex (15), we have constructed a reliable docking model of the aRF1·aEF1α·GTP complex, to elucidate the structural basis for the GTPase-mediated mechanism of stop codon decoding via tRNA mimicry. The biological implications for the evolution of the tRNA mimicry systems are discussed.
Fig. 1.
Conservation of the putative GTPase binding domain site 1 of aRF1. (A) Schematic presentation of eRF1 and two major groups of aRF1s (Euryarchaeota and Crenarchaeota). As a member of Crenarchaeota, A. pernix aRF1 lacks a large portion of domain C (“Δ”). Site 1, the predominant GTPase binding site found in the eRF1·eRF3-d23 crystal structure (located separately in sequences 1A and 1B) (10), and other relevant sites are indicated. The numbers of amino acid residues are indicated for representatives of each group in brackets. Homologous Pelota regions (domains B and C) are indicated in red. (B) Alignment of representative aRF1/eRF1/Pelota amino acid sequences around site 1 in A. The black dots above the sequences indicate the previously demonstrated critical contact residues for eRF3 binding (10). Distances in the eRF1·eRF3-d23 complex (4 Å, 6 Å) are also indicated in colored dots at the top. (C) Superposition of the domain C (site 1) structures of aRF1 and eRF1. The extra region of eRF1, which is truncated in the Crenarchaeota aRF1 group, is indicated as Δ. This is a rear side view of Fig. S3A.
Results and Discussion
Atomic Structure of A. pernix aRF1.
The crystal structure of an archaeal class-I release factor (aRF1) from A. pernix (16), an extremophilic Crenarchaeon, was determined by the multiple-wavelength anomalous dispersion method and refined to 2.2-Å resolution (Tables S1 and S2). The overall structure of aRF1 is organized into three domains, domains A, B, and C (Fig. S3A). As expected from the sequence similarity between aRF1 and eRF1, each of the three domains of aRF1 has a structure similar to the corresponding domain of eRF1 (17) (rmsd values of 1.2 Å, 1.6 Å, and 1.9 Å, for domains A, B, and C, respectively). However, the domain arrangement is completely different from that of eRF1 (see below for details). In addition, the loops formed by residues 100–104 in domain A and 175–186 in domain B of aRF1 are disordered in the crystal structure and thus were not modeled.
Structural Conservation of the Putative GTPase Binding Site in aRF1 and aPelota.
In the process of translation termination in eukaryotes, eRF1 is bound to eRF3 and thereby delivered to the A site of the ribosome presenting the stop codon. The conserved C-terminal part of eRF3 (18) consists of three domains (domains 1, 2, and 3), which share structural similarity to the translational GTPases, including the eukaryotic and archaeal EF1α proteins. The crystal structures of eRF1 in complex with domains 2 and 3 of eRF3 (eRF3-d23) have been reported, in which domain C of eRF1 forms hydrophobic interactions with domain 3 of eRF3, constituting the primary binding site (10) (site 1).
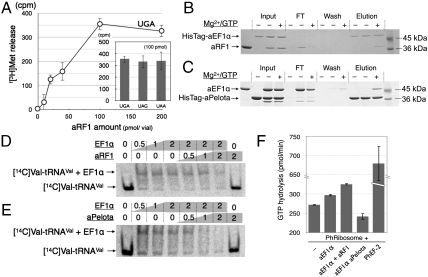
The aRF1 from Aeropyrum pernix (ape-aRF1) has a large deletion in the middle of domain C, as a hallmark of this phylum (Fig. 1A). This truncation, along with the distinctive arrangements between domains B and C (described later), causes the overall structure of aRF1 to be strikingly compact, as compared to the apo- and semicomplexed structures of eRF1 (PDB ID codes 1DT9 and 3E1Y, respectively), and it fits the shape of the tRNA well (Fig. S3B). Despite its simple structure, the purified recombinant aRF1 exhibits polypeptide-chain releasing activity in response to each of the three stop codons programmed on the mammalian 80S ribosome (Fig. 2A). Moreover, the residues involved in the site 1 interaction are well conserved in aRF1 (Fig. 1B) and maintain the domain structure (Fig. 1C; also see Fig. 3).
Fig. 2.
In vitro analysis of aRF1 and Pelota for aEF1α binding. (A) The f[3H]Met release assay of aRF1, using HeLa S3 ribosomes programmed with a stop-codon containing mini mRNA. The dose curve for UGA-dependent release and the activities at 100 pmol/50-μL reaction volume are shown in the histogram. The in vitro binding of HisTagged-aEF1α to aRF1 (B) and the in vitro binding of HisTagged-aPelota to aEF1α (C) are demonstrated by pull-down assays. The reaction mixtures including the factors [“Input,” HisTagged protein only (Left), HisTagged protein and nontagged protein (Middle), both with Mg2+/GTP (Right)] were mixed with MagneHis™ Ni particles and the HisTagged proteins were immobilized, and then the HisTagged proteins and their binding factors were resolved by SDS-PAGE. The flow-through fraction (FT), the wash fractions (Wash), and the eluted fractions (“Elution”) are shown. In vitro competition assays for the aEF1α-[14C]Valyl-tRNAVal complex by aRF1 (D) and aPelota (E) are shown. The mixtures of [14C]Valyl-tRNAVal with the factors indicated above were resolved by native PAGE, and the shifted bands were analyzed. (F) GTPase assay with Pyrococcus ribosomes. The ribosomes were mixed with various combinations of aEF1α (10 pmol), aRF1 (40 pmol), and aPelota (40 pmol), or with Ph EF-2 (10 pmol, as a control of GTP hydrolysis by translational GTPase in Pyrococcus horikoshii ribosomes), as indicated. See Materials and Methods for details.
Fig. 3.
aRF1·aEF1α·GTP complex model and interactions between aRF1 and aEF1α. (A) A docking model of the aRF1·aEF1α·GTP complex. aEF1α is colored red (domain 1), orange (domain 2), and yellow (domain 3); aRF1 is colored as in Fig. S3. The bound GTP is shown in a green-colored stick model. The Mg2+ is shown as a magenta sphere. The five interaction sites between aRF1 and aEF1α are indicated, and A–D for domain B of aRF1 (red), and site 1 for domain C of aRF1 (green) are boxed. Domain B (aRF1) interactions with aEF1α are observed at four sites: A, switch I region; B, switch II region; C, domain 2 cleft region; D, domain C linker helix region. (B) The interaction sites are shown with the relevant amino acid residues depicted by stick models, colored red for those of aEF1α, and black for aRF1. The GGQ motif is modeled according to human eRF1 structure. Figures were rendered using Pymol (32).
Recently, the apo-form crystal structures of an aRF1/eRF1-related protein, Pelota (Dom34p in budding yeast), from both a eukaryote and an archaeon were reported (12, 13). Pelota binds another EF1α family protein, Hbs1, and is considered to play a crucial role in mRNA surveillance for protein synthesis, such as in No-go decay (NGD), in which endonucleolytic cleavage of the mRNA is induced upon translation arrest, in order to eliminate the aberrant mRNAs and rescue the stalled ribosomes (14). Although little is known about the function of Pelota, the structural similarities to eRF1 and aRF1 are predominantly observed in domains B and C, including site 1 (Fig. 1B).
Based on these structural similarities, aRF1 and archaeal Pelota (aPelota) are assumed to bind to a putative translational GTPase to be delivered to the ribosome. However, in spite of the phylogenetic conservation of eRF1/aRF1 and Pelota/aPelota in eukaryotes and archaea, no archaeal orthologs of eRF3 and Hbs1 have been identified by genome analyses. Because the only EF1α family GTPase found in archaeal genomes is aEF1α (8), it is tempting to hypothesize that aEF1α plays multiple roles as the EF1α-like GTPase for both aRF1 and aPelota, as well as for the canonical tRNAs. Domains B and C of aRF1 and aPelota share higher sequence similarity than those of eRF1 and Pelota, thus supporting this hypothesis.
Multiple Functional Binding of ape-EF1α to aRF1 and aPelota.
In vitro pull-down experiments using tagged and nontagged recombinant proteins were performed, to examine the binding capability between aEF1α and aRF1/aPelota. aRF1 was copurified with His-tagged aEF1α only in the presence of Mg2+/GTP (Fig. 2B). Similarly, aEF1α was copurified with His-tagged aPelota only in the presence of Mg2+/GTP (Fig. 2C). These results clearly indicate that the GTP form of archaeal aEF1α binds both aRF1 and aPelota. The yeast two-hybrid analysis also demonstrated the binding of aRF1 and aPelota to aEF1α (Fig. 4A).
Fig. 4.
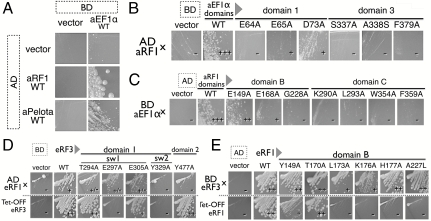
Yeast two-hybrid analysis of aRF1/Pelota binding to aEF1α at sites 1 and 2. (A) Two-hybrid analysis (5-d growth) between wild-type aEF1α (binding domain, BD) and aRF1/aPelota (activation domain, AD). (B) Two-hybrid analysis (3-d growth) of mutants on site 1 and site 2 residues in aEF1α (BD) against aRF1 wild type (AD). (C) Two-hybrid analysis (3-d growth) on mutants of site 1 and site 2 residues in aRF1 (AD) against aEF1α wild type (BD). (D) Mutational analysis of site 2 residues in Sp-eRF3. The upper row presents the results of the two-hybrid interaction of Sp-eRF3 mutants (BD) against Sp-eRF1ΔN2-F288A (AD). The lower row presents the results of the complementation of Tet-off eRF3 S. cerevisiae strain. (E) Mutational analysis on site 2 residues in SP-eRF1. The upper row presents the results of the two-hybrid interaction of Sp-eRF1 mutants (AD) against Sp-eRF3-Y577A (BD). The lower row presents the results of the complementation of Tet-off eRF1 S. cerevisiae strain.
Because the predicted interface of eRF3 for binding with eRF1 overlaps with the interface of EF1α for binding aminoacylated tRNAs, as deduced from tRNA·EF-Tu·GTP ternary complex structures (19), aRF1, aPelota, and the aminoacylated tRNA are expected to bind exclusively to EF1α. The competitive binding of aRF1 and aPelota to the aEF1α·tRNA·GTP complex, i.e., aEF1α complexed with valyl-tRNAval in the presence of GTP, was examined and indicated that both aRF1 and aPelota bind to EF1α competitively, replacing the aa-tRNA (Fig. 2 D and E).
The ribosome-dependent GTPase assay of EF1α, using the thermostable ribosome from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 (20), demonstrated the aRF1-dependent GTPase activation of EF1α (Fig. 2F). This clearly indicates that aRF1 forms the ternary complex with aEF1α to promote ribosome binding in the presence of GTP for the stop codon dependent peptide-chain release (Fig. 2A), as demonstrated in eRF1/eRF3 cases (11). On the other hand, aPelota strongly inhibited the GTPase activity of aEF1α to a level even lower than that of the residual GTPase background accompanying the Pyrococcus ribosome, probably due to its tight and stable complex formation upon binding to the ribosome (Fig. 2F). Although the physiological significances of much slower GTP hydrolysis rate exhibited by aPelota·aEF1α·GTP complex with free Pyrococcus ribosome is not clear at present, this result suggests that aPelota form a complex with aEF1α prior to ribosomal entry in the presence of GTP. The structural interpretation based on the aPelota·aEF1α·GTP ternary complex structure, in which the prolonged stall of the aPelota·aEF1α·GTP complex at the ribosomal A site by slow GTP hydrolysis promotes mRNA decay pathway (15), could be worth further analysis. There is, unfortunately, no feasible experimental system to analyze overall activities of archaeal aRF1 and aPelota in translation termination and mRNA surveillance; however, our results strongly indicate the functional coupling of the complex formation of aRF1 and aPelota with aEF1α.
Modeling of the aRF1·aEF1α·GTP Complex for Prediction of the Regulatory Contact Sites Between aRF1 and aEF1α.
The present in vivo and in vitro analyses demonstrated that aEF1α interacts with aRF1 and Pelota (aPelota), as well as the aa-tRNA. To elucidate the molecular details of these interactions, we recently solved the crystal structure of the ternary complex of A. pernix aPelota, aEF1α, and GTP [Fig. 5, Middle (15)]. The overall shape is strikingly similar to that of the tRNA·EF-Tu·GTP complex structure. Moreover, the first atomic structure of the aPelota·aEF1α complex revealed the detailed interactions between aEF1α and aPelota, in which domains B and C of aPelota are recognized by aEF1α in the GTP-bound form. Domain 3 of aEF1α and domain C of aPelota form a quite similar interaction to that observed at site 1 of the eRF1·eRF3-d23 complex (10). However, the linker helices connecting domains B and C of aPelota in the Pelota·aEF1α complex exhibit a sharply bent conformation mediated by hydrophobic interactions (15), facilitating the concomitant binding of domains B and C to aEF1α. In contrast, the conserved linker helices in the previous eRF1 (Fig. S4C, Right) and Pelota/Dom34 structures are stretched, and thus the two putative concomitant binding surfaces are sequestered; e.g., when site 1 is fixed, domain B of eRF1 in the eRF1·eRF3-d23 complex (PDB ID code 1DT9) is rotated by 114° relative to domain B of aRF1 (Fig. S4C).
Fig. 5.
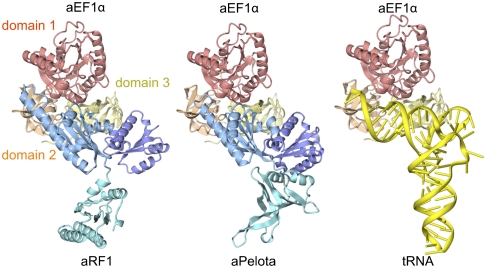
tRNA molecular mimicry in translation and structural basis for GTPase-mediated stop codon decoding. The complex models for aRF1·aEF1α·GTP (Left) and aa-tRNA·aEF1α·GTP (Right) are exhibited with the aPelota·aEF1α·GTP complex structure (15).
Interestingly, the overall structure, including the orientations of domains B and C of aRF1, is quite similar to that of aPelota bound to aEF1α. Moreover, the linker helices of aRF1 also adopt a bent conformation (Fig. S4), which is stabilized by hydrophobic interactions between domains B and C, involving L158, L163, V267, and M268 of domain B, and Y279 and V283 of domain C (Fig. S4 A and B). Therefore, aRF1 domains B and C in the present structure can be directly docked onto the GTP-bound form of the aEF1α surface (Fig. S5), in a similar manner to the aPelota·aEF1α complex. Hence, we constructed a reliable docking model of the aRF1·aEF1α·GTP complex based on the sequence and structural similarity (Fig. 3A).
The surface groove of domain C of aRF1, which interacts with domain 3 of aEF1α, is composed mainly of hydrophobic amino acids. These residues are located in helices α9 and α12, and strands β9 and β12, and form a hydrophobic patch. The interaction manner between domain C and domain 3 of the aRF1· aEF1α·GTP complex seems to be essentially identical to that of the site 1 interactions of the eRF1·eRF3·GTP complex described above. F359 of aRF1 and Y343 of aEF1α are highly conserved, and the corresponding residues were previously shown to be important for the interaction between eRF1 and eRF3 of S. pombe (site 1 in Fig. 3 A and B). In our yeast two-hybrid assay, the aRF1 protein with the mutations of residues K290, L293, W354, and F359, corresponding to F288, I291, Q400, and F405 of eRF1, respectively, showed a significant reduction in aEF1α binding (Fig. 4C). Consistently, the mutation of aEF1α at residues S337, A338, and F379, corresponding to S571, I572, and F612 of eRF3, showed a weaker interaction with aRF1 (Fig. 4B, domain 3; Fig. 4C, domain C). These results are also consistent with the previous analysis of the eRF1-eRF3 interaction at site 1 (10).
Interaction Sites by Domain B of aRF1 Bridging All Structural Domains of aEF1α.
In the current aRF1·aEF1α·GTP complex, domain B of aRF1 seems to interact with all of the structural domains of aEF1α in essentially the same manner as aPelota (15), through conserved interaction sites. The interaction sites in the aRF1·aEF1α·GTP complex model were examined for aRF1/aEF1α as well as eRF1/eRF3 by yeast two-hybrid binding assays and/or yeast growth complementation assays, as follows.
Switch Regions I and II of Domain 1.
The side chain of the strongly conserved K176, on the loop between β7 and α5 of aRF1, interacts with the side chains of E65 and T75, and the main chain oxygen of D73 in the switch I region of aEF1α (“A,” Fig. 3B). Alanine substitutions at E65 and D73 of aEF1α mildly reduced the two-hybrid binding with aRF1 and, consistently, corresponding substitutions at E297 and E305 of S. pombe (hereafter referred to as Sp-eRF3) also reduced the two-hybrid binding with S. pombe eRF1. Mutations of the residues E64A of aEF1α, G228A of aRF1, and T294A of Sp-eRF3 (M62 in aEF1α), which were estimated to be crucial for this binding site inferred from the aPelota complex, drastically reduced the two-hybrid binding (Fig. 4 B–D).
The region connecting strand β5 to β6 of aRF1 consists mainly of acidic residues, in which the side chains of E149 and D151 of aRF1 form salt bridges to K99 in the aEF1α switch II region (“B,” Fig. 3B). In addition, the highly conserved P227 and G228 residues, on the loop between β8 and α6, contribute to the formation of the small convex area of aRF1, which contacts F97 of aEF1α, and exhibits a shape complementary to that of the switch I and II regions of aEF1α. Supporting this proposal, the mutations of E149A and P227A in aRF1, as well as Y329A in Sp-eRF3 (F97 in aEF1α), reduced the aRF1-aEF1α interaction in a yeast two-hybrid assay (Fig. 4 B–D).
Domain 2.
The residues of β-strand a2 and the loop region between β-strand e2 and β-strand f2 form a narrow cleft with the switch region I of domain 1 of aEF1α (Fig. S2), which would sandwich the loop region between the β-strand β7 and the helix α7 of aRF1, including the GGQ motif (“C,” Fig. 3B). Mutations in this region, such as Y477A of Sp-eRF3 (Y241 in aEF1α), reduced the yeast two-hybrid binding (Fig. 4D), and the mutants L173A and K176A of S. pombe eRF1 (hereafter Sp-eRF1) (V173 and K176 of aRF1) (Fig. 4E) exhibited functional defects in a growth complementation assay. Importantly, the H177A Sp-eRF1 mutation (H177A of aRF1) resulted in a strong dominant negative effect for cell growth and translation termination in the budding yeast, suggesting that the H177 residue of aRF1 is crucial for regulatory interactions.
Domain 3.
K394 of aEF1α and E168 of aRF1, as well as D414 of aEF1α and K269 of aRF1, the latter of which resides in the linker helix region (α8, α9) of aRF1, form close contacts in the model. Previously, we reported that the D647A mutation in Sp-eRF3 (D414 in aEF1α) reduced its binding to Sp-eRF1 (10). Consistent with this observation, the E168A mutation in aRF1 also reduced its binding to aEF1α (Fig. 4C).
Consequently, our analyses clearly demonstrated the functionality of all of the contact sites in aRF1/aEF1α as well as eRF1/eRF3 suggested by the current aRF1·aEF1α·GTP complex model. Despite their overall similarities, the docking orientation of domain B vs. domain C of aRF1 largely differs from that of eRF1 in our previous eRF1·eRF3·GTP complex model (10), which was constructed by simple rigid-body transformation of the eRF1·eRF3-d23 crystal structure to fit the lower resolution image of the eRF1·eRF3·GTP complex revealed by a SAXS experiment. The mild defects shown by the alanine substitutions at residues 189 and 200 of eRF1 (in S. pombe; 192 and 203 in human), which protrude from the complex and lack contacts with eRF3 in the current model (in “C” of Fig. 3B) and were proposed to be involved in GTPase activation in our previous study, now suggest that those conserved residues are rather important for other interactions, such as ribosomal binding and/or the translocation of domain B in the A site. The current aRF1·aEF1α·GTP complex model described in this study updates the previous model and sheds light on the details of the interactions between aRF1/eRF1 and aEF1α/eRF3 and also provides a functional interpretation of the GTPase in translation termination (depicted in Fig. S6). A summary of the contact site residues in this study is provided in Table S3.
Biological Implications for the Multiple Roles of aEF1α for tRNA and tRNA Mimicry Proteins.
We constructed a model of the tRNA-aEF1α complex, based on the tRNA·EF-Tu ·GTP complex structure. The overall structures of the aRF1·aEF1α·GTP, aPelota·aEF1α·GTP, and tRNA·aEF1α·GTP complexes are very similar (Fig. 5). In the structure of the aPelota·aEF1α·GTP complex, a negatively charged patch is formed on the surface of aPelota domain B, which is recognized by K99 of aEF1α. A similar acidic patch is also formed by E149, D151, and E168 of aRF1 domain B, which may also be recognized by K99 of aEF1α. Moreover, the residues constituting the acidic patch coincide well with the phosphate groups at positions 1, 2, and 67 in the tRNA acceptor stem. In the structure of the Thermus aquaticus aa-tRNA·EF-Tu complex, these phosphate groups are recognized by K90 of EF-Tu (19), which corresponds to K99 of aEF1α and is widely conserved among the aEF1α/EF1α/EF-Tu subfamilies. Therefore, the acidic patch conserved between aRF1 and aPelota may enable their binding to aEF1α, by mimicking the tRNA phosphate backbone.
Extensive phylogenetic analyses have revealed the absences of eRF3 and Hbs1 orthologs in archaeal genomes and have shown that the eRF3 and Hbs1 proteins are highly conserved throughout eukaryotic species, whereas the authentic Hbs1 is missing in a few eukaryotic Apicomplexa species, despite the presence of apparent Pelota/Dom34 orthologs in the genome (8). Thus, it was originally proposed that GTPase participation is dispensable in the archaeal and/or some primitive branches of eukaryotes for NGD as well as for translation termination (8). Those situations might be partially explained by the fact that Hbs1 is nonessential, at least for the growth of budding yeast (21), and the fact that eRF1/aRF1 (6, 7) or the simple ape-aRF1 per se can catalyze peptide-chain release in vitro without GTPase counterpart molecules, as in the bacterial class-I RFs. However, our present results strongly suggest that the tRNA mimicking proteins, aRF1/eRF1 and aPelota/Pelota, are tightly coupled with the translational GTPase activities and coevolved from a cenancestor of eukaryotes and archaea.
Similarly, it has been reported recently that the pair of elongation factor G (EF-G)-related GTPases, EF-G1 and EF-G2, exclusively catalyze either translocation or ribosome recycling reactions in human mitochondria, whereas in most bacterial species a single authentic EF-G catalyzes both reactions (22). Those facts tell us certain aspects of differentiation in the evolution of universal translational GTPases families, EF1α, eRF3, and Hbs1. The genes encoding translational GTPases are often present in multiple copies in the genomes of many organisms, presumably to support efficient protein synthesis. It is well known that translation termination as well as mRNA surveillance systems recruit many additional regulatory factors and are incorporated into various regulatory networks, especially in higher organisms (8). Therefore, it is tempting to assume that gene multiplication of a cenancestral GTPase for an omnipotent EF1α might have triggered the functional compartmentation of the regulatory mechanisms in individual tRNA mimicking systems (schematized in Fig. S7) (23).
Materials and Methods
Expression and Purification of A. pernix aRF1.
The gene encoding full-length A. pernix aRF1 was cloned into the pET28c vector (Novagen) and was overexpressed in the Escherichia coli BL21(DE3)RIL strain. SeMet-labeled aRF1 was expressed in the B834(DE3)RIL strain.
Crystallization and Data Collection.
SeMet-labeled aRF1 was concentrated by ultrafiltration to 5.3 mg/mL. Crystals of SeMet-labeled aRF1 were grown at 20 °C, by mixing equal amounts of the protein solution (4.8 mg/mL aRF1 in buffer E containing 10 mM ATP) and the reservoir solution (200 mM ammonium citrate tribasic, pH 7.0, 12% PEG monomethyl ether (MME) 2000, 10 mM ATP). Crystals were cryoprotected in 25% ethylene glycol, 220 mM ammonium citrate, pH 7.0, 14% PEG MME 2000, and 11 mM ATP, and were flash-cooled at 100 K. Native crystals were grown from seed crystals of SeMet-labeled aRF1, cryoprotected, and flash-cooled under the same conditions as described above. Diffraction data were collected at the beamlines BL41XU at SPring-8 and NW12A at KEK PF-AR and were processed with the program HKL2000 (HKL Research). The statistics of the data collection are summarized in Table S1. The structure factors and coordinates of aRF1 have been deposited in the Protein Data Bank (PDB ID code 3AGK).
Structure Determination and Refinement.
The structure was determined by the multiwavelength anomalous dispersion method, using the selenium anomalous signal. Twelve of the 14 selenium sites were detected by the programs SHELXC and SHELXD (24), with the dataset up to 2.8 Å collected at the peak wavelength. Heavy-atom refinement and phase calculations were performed with the program SHARP (25), followed by solvent flattening with the program SOLOMON. The automatic model building was performed with the program RESOLVE. The model was subsequently improved through alternate cycles of manual building using the program COOT (26) and refinement with the program PHENIX (27), after 5% of the reflections had been set aside to calculate the Rfree. The final model was refined against the native dataset up to 2.2 Å to an Rwork of 23.8% with an Rfree of 29.3%.
Release Assay of aRF1.
The stop codon dependent f[3H]Met release assay was performed as described previously (28), using 80S ribosomes purified from the HeLa S3 cell line. The concentration of aRF1 is indicated as pmol per 50-μL reaction volume. The amount of f[3H]Met released at zero time was subtracted from all values. Assay conditions and other factor preparations were described previously (29).
Preparation of P. horikoshii ribosomes and GTPase assay.
Archaeal ribosomes were prepared from P. horikoshii cells as described previously (28), except that an extraction buffer containing 20 mM MgCl2, 10 mM KCl, 5 mM 2-mercaptoethanol, and 20 mM Tris-HCl, pH 7.6, was used. The GTPase activity dependent on the factors was assayed in the presence of 2.5 pmol of P. horikoshii ribosomes, in a solution (20 μL) containing 30 nmol [γ-32P]GTP, 7 mM MgCl2, 50 mM NH4Cl, and 20 mM Tris-HCl, pH 7.6 After an incubation at 90 °C for 10 min, the liberated inorganic phosphate was extracted and quantitated, as described previously (30).
Pull-Down and tRNA Competition Gel Shift Assays.
Each of the HexaHis-tagged and tag-cleaved purified proteins was mixed (at molar ratios of aEF1α∶aRF1 = 1∶1, aEF1α∶aPelota = 2∶1) and incubated in the binding buffer (100 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM MgSO4, 1 mM GTP) at 70 °C for 1 min, and then the His-tagged protein was immobilized with MagneHis™ Ni particles. The beads were washed twice with the binding buffer containing 1 mM imidazole, and then the bound proteins were copurified with the binding buffer containing 500 mM imidazole. The fractions were analyzed by 12.5% SDS-PAGE. For the tRNA competition assay, aRF1, Pelota, and the valyl-tRNAval transcript, charged with [14C]valine by the valyl-tRNA synthetases [NP_148179.2], were mixed at the indicated molar ratios and incubated with EF1α at room temperature for 5 min. The reaction mixtures were then analyzed on a native acrylamide gel, and the image was analyzed with a BAS3000 image analyzer system (Fuji Film Co., Ltd.).
Yeast Two-Hybrid Assays and Growth Complementation Assays.
For the two-hybrid assays, the genes encoding the wild-type and mutated aRF1 were constructed in the AD vector. The eRF1 binding variants were constructed on the N-terminal truncated form of eRF1 (i.e., Sp-eRF1ΔN2), with efficient binding capacity for eRF3, in the AD vector. For the BD vectors, the wild-type and mutated aEF1α or eRF3c (residues 196–662 of S. pombe) sequences were cloned into the pGBT9 vector (Clontech). The in vivo two-hybrid assay was performed with the S. cerevisiae AH109 strain, using the same procedures and conditions as described previously (31).
For growth complementation, the wild-type and mutated eRF1 and eRF3 sequences were cloned into p416ADH plasmid (10). The growth of the tetracycline repressible eRF1 and eRF3 strains, tet-OFF sup45 and tet-OFF sup35, transformed by the eRF1 and eRF3 expression plasmids, respectively, was examined on agar plates with 7.5 μg/mL of doxycycline.
Supplementary Material
Acknowledgments.
We thank Prof. Y. Nakamura for general support of this project, RIKEN BioResource Center (Ibaraki, Japan) for providing genomic DNA of Aeropyrum pernix, M. Kimura for the kind gift of the P. horikoshii cell preparation, T. Honda for the preparation of ribosomes from P. horikoshii, and the beamline staffs at NW12A and NE3A of KEK PF-AR and BL41XU of SPring-8 for assistance in data collection. This work was supported by a PRESTO Program grant from Japan Science and Technology (JST) (to K.I.), by a Strategic International Cooperative Program grant from JST (to O.N.), by a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to O.N.), by grants from MEXT (to K.I., R.I., and O.N.), and by grants from The Uehara Memorial Foundation (to O.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3AGK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009599107/-/DCSupplemental.
References
- 1.Nakamura Y, Ito K. Making sense of mimic in translation termination. Trends Biochem Sci. 2003;28:99–105. doi: 10.1016/S0968-0004(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 2.Ito K, et al. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 3.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–20. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–827. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 5.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dontsova M, et al. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 7.Frolova L, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson GC, et al. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolova LY, et al. Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Z, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Graille M, et al. Structure of yeast Dom34: A protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 14.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K, et al. Structural basis for ribosomal mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc Natl Acad Sci USA. 107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawarabayashi Y, et al. Complete genome sequence of an aerobic hyper-thermophilic Crenarchaeon, Aeropyrum pernix Kl. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 17.Song H, et al. The crystal structure of human eukaryotic release factor eRF1- mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 18.Kong C, et al. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 19.Nissen P, et al. Crystal-structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 20.Kawarabayashi Y, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 21.Nelson RJ, et al. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 22.Tsuboi M, et al. EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol Cell. 2009;35:502–510. doi: 10.1016/j.molcel.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 23.Long M, et al. The origin of new genes: Glimpses from the young and old. Nature Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 24.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 25.de La Fortelle E, Bricogne G. Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Maone E, et al. Functional analysis of the translation factor aIF2/5B in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2007;65:700–713. doi: 10.1111/j.1365-2958.2007.05820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tate WP, Caskey CT. Termination of protein synthesis. In: Spedding G, editor. Ribosomes and Protein Synthesis: A Practical Approach. Oxford, England: IRL Press at Oxford Univ Press; 1990. pp. 81–100. [Google Scholar]
- 30.Frolova L, et al. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 31.Ito K, et al. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.