Abstract
Methylation of cytosine in the 5 position of the pyrimidine ring is a major modification of the DNA in most organisms. In eukaryotes, the distribution and number of 5-methylcytosines (5mC) along the DNA is heritable but can also change with the developmental state of the cell and as a response to modifications of the environment. While DNA methylation probably has a number of functions, scientific interest has recently focused on the gene silencing effect methylation can have in eukaryotic cells. In particular, the discovery of changes in the methylation level during cancer development has increased the interest in this field. In the past, a vast amount of data has been generated with different levels of resolution ranging from 5mC content of total DNA to the methylation status of single nucleotides. We present here a database for DNA methylation data that attempts to unify these results in a common resource. The database is accessible via WWW (http://www.methdb.de). It stores information about the origin of the investigated sample and the experimental procedure, and contains the DNA methylation data. Query masks allow for searching for 5mC content, species, tissue, gene, sex, phenotype, sequence ID and DNA type. The output lists all available information including the relative gene expression level. DNA methylation patterns and methylation profiles are shown both as a graphical representation and as G/A/T/C/5mC-sequences or tables with sequence positions and methylation levels, respectively.
INTRODUCTION
In plants, vertebrates and certain fungi, DNA methylation is related to a number of epigenetic (i.e. heritable, but potentially reversible) phenomena (1). In vertebrates, the DNA methylation pattern is established early in embryonic development and in general the distribution of 5-methylcytosine (5mC) along the chromosomes is maintained during the life span of the organism (for review see 2). Methylation in particular regions of genes or in their vicinity can inhibit the expression of these gene (3). Artificial demethylation of genes has been shown to result in reactivation. Recent work has provided evidence that the gene silencing effect of methylated regions is accomplished through the interaction of methylcytosine binding proteins with other structural compounds of the chromatin (for review see 4). This interaction probably makes the DNA inaccessible to transcription factors through histone deacetylation and chromatin structure changes (for review see 5). Differentially methylated regions are also key elements in the transcriptional regulation of genes whose expression state depends on the sex of the parent from whom they were inherited (imprinted genes) (for review see 6). The initiation and the maintenance of the inactive X-chromosome in female eutherians was found to depend on methylation (for review see 7). Changes in the methylation patterns seem to occur during developmental and pathologic changes of cells. In particular, tumor tissue of mammalia was found to be characterized by a genome wide demethylation and a local hypermethylation of tumor-suppressor genes (for review see 8).
Despite a long history of DNA methylation studies and in spite of the increasing interest in DNA methylation patterns, so far no attempt has been made to collect and store the available methylation data in a public database. When DNA methylation patterns are published, the authors display them usually in a graphical way. This is convenient for pattern recognition and data representation, however it makes it difficult, if not impossible, to compare patterns from different sources or to analyze the data with appropriate software tools. The common bibliographic search engines will only deliver information that is contained in the title, the abstract and the keywords of a publication but they will fail when a systematic survey about the available data is necessary. For this reason we established a new database for DNA methylation patterns and propose to store all available data about DNA methylation in this database.
Catalogs and other forms of databases have always been indispensable tools for biologists (for review see 9). Nowadays the World Wide Web provides a simple way to make databases accessible to the public. ‘MethDB’, the DNA methylation database described in this paper, stores data about the degree of methylation in total DNA, subfractions of total DNA, DNA fragments and single nucleotide positions. It contains information about the nature and the origin of the investigated sample and data about the experiments. Methylation patterns and profiles are represented in a graphical and alphanumeric way. A particular objective of this database is to unify the heterogeneous data about DNA methylation into one common resource and to cross-relate the entries to other existing databases via HTML links.
Structure of the database and file formats
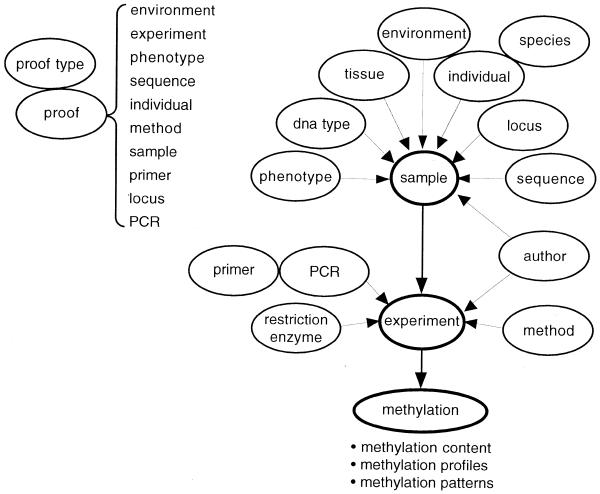
MethDB was designed to store heterogeneous data from different kinds of experiments. The database system has an open structure which consists of 18 tables (Table 1) and can easily be extended. The tables are related to each other (Fig. 1). The three central tables are ‘sample’, ‘experiment’ and ‘methylation’. A ‘sample’ record contains the description of the object, or target, of an experiment. In ‘experiment’ the experimental conditions and procedures are stored. Finally, ‘methylation’ contains the results of the experiments. For the moment we distinguish three sorts of results: methylation content, methylation profile and methylation pattern. Methylation content is the relative share of 5mC in a DNA sample without having information about the position of 5mC in the sequence. A methylation profile consists of a series of values for the methylation level at cytosine positions along a known sequence. Methylation patterns describe the sequence of the five nucleotide bases G,A,T,C and 5mC in individual DNA molecules. DNA sequences, methylation profiles and methylation patterns are stored in flat ASCII format.
Table 1. The different tables of MethDB.
| Table name |
Purpose |
| Author | Contains information about how the person who has submitted an entry can be contacted (name, address, telephone number and email address). |
| Proof type | A list of reference types. A proof type can for instance be an URL, a GenBank ID or an OMIM ID. |
| Proof | Each MethDB entry must be provided with a proof, i.e. a direct or indirect reference to a source where the original data can be found. In particular, this table is useful to address the URL of database cross-references. |
| DNA type | Lists DNA types (e.g. single genomic fragment, low repetitive sequence, total DNA) and contains short descriptions of the DNA types. |
| Phenotype | Describes the phenotype of the investigated specimen. By convention, tumors are given the phenotype name ‘tumor –’ followed by its designation. Each phenotype is described in brief and if possible a reference is provided. |
| Species | Table of species names in scientific and trivial notation. |
| Tissue | Provides tissue names and short descriptions of the tissues. |
| Locus | The fields of this table are locus, species, chromosome, mapping location, gene product, proof and comment. It contains the information about defined pieces of DNA. These DNA fragments can be, for instance, genes or unique hybridization loci. ‘locus’ is the unique key. In case a gene is described, ‘locus’ is identical to the GenBank name of the gene (/gene=”” in the GenBank feature list). |
| Method | Contains the name of the method that was used to generate the methylation data, its description and the reference. |
| Primer | Stores the sequences of primers, their target sequences (which are not necessarily the same) and the reference for the primer. |
| PCR | If PCR was used in the experiment to obtain the methylation data, this table stores the information about the used primers, the PCR conditions and the reference for this PCR. |
| Restriction enzyme | Contains information about restriction enzymes (name, recognition sites and methylation sensitivity). It is intended to establish a ‘clickable’ link from this table to the restriction enzyme database REBASE (http://rebase.neb.com). |
| Sequence | Stores the unique sequence ID, the name of the file that contains the sequence in FASTA format, the associated gene or locus, the reference for the sequence, a comment and the 5′-, 3′-neighboring and complement strands, as far as they exist. The sequence itself is not contained in the database but exists as a file (‘sequence_sequence ID’). |
| Individual | Contains data about the investigated individual. Each individual is given a unique lifelong individual ID. An arbitrary name, the species, the strain, the sex and the reference can be entered. |
| Environment | Environmental impact on the investigated sample. Currently the main purpose of this table is to provide information about the nutrition of the investigated individual. |
| Sample | Unifies all information about the investigated DNA under a unique sample ID, its origin (gene, sequence, individual, tissue) and the developmental state, age and phenotype of the individual the sample was taken from. It also contains the gene expression data if a particular gene was studied. For each sample ID one or more than one experiment can exist. |
| Experiment | Contains the information about the experiment: its ID and the sample ID of the sample that was used for the experiment, the type of DNA that was investigated and the method that was applied. If PCR was used the PCR ID is listed and if restriction enzymes were used they are indicated. The results of the experiment are stored either directly in this table (in case a single value like mol% 5mC was obtained) or in the related table ‘methylation’ if more than one value was obtained. This is always the case when a particular sequence was studied and the methylation at each or at least a number of cytosine positions was determined. |
| Methylation | Stores the file name of the file that describes the methylation of distinct cytosines in a DNA sequence. The table is connected with ‘experiment’ by a 1:n relation. One experiment can deliver more than one description of methylation patterns, i.e. multiple entries in the ‘methylation’ table. The table contains an ID for each entry, the ID of the experiment, the information if single or multiple DNA strands were investigated, if it was single strand or double strand DNA that was studied and the file names of the files that describe the methylation profile or methylation patterns, respectively. |
The methylation database consists of 18 individual tables that are interrelated.
Figure 1.
The structure of MethDB. Each circular element represents one table in the database system. Arrows describe 1:n relationships in the direction of the arrowhead. On one sample, or experimental target, more than one experiment can be performed. Each experiment can deliver several methylation patterns but only one methylation profile or one methylation content value. For the sake of clarity of the figure, the relation of the proof table to the other tables is displayed separately, on the upper left corner of the figure.
Sequences. Sequences are simple text files in FASTA format (>name[new line]sequence[new line]) (10). The name is the sequence ID. These files contain the investigated sequences in the standard IUB notation. File-name is ‘sequence_sequence ID’. An example is shown in Figure 2a.
Figure 2.
Examples of the different file formats used in MethDB. (a) A sequence file in FASTA format. (b) An example for the format of the tables that are used to describe methylation profiles. (c) An example for the modified FASTA format that is used to store methylation patterns of single molecules: 5mC is indicated as upper case ‘C’ while all other nucleotide bases are written in lower case letters.
For methylation profiles and patterns we introduce two new file formats:
Methylation profiles. Methylation profiles are described by tables of two comma-separated values: position in a mother-sequence and degree of methylation at this site between 0 (no methylation) and 1 (complete methylation). ‘na’ instead of the methylation level indicates that the position was not analyzed and that this fact has been mentioned in the original publication. Each pair of position/methylation-level values is followed by a [new line] character. The first line of the table contains the symbol ‘>’ followed by the metpos ID. The table is separated from an optional comment by two slashes ‘//’. File-name is ‘methpostbl_metpos ID’. Figure 2b shows an example table.
Methylation patterns. Methylation patterns are sequence files in FASTA format with all nucleotide bases, except 5mC, written in lower case and 5-mC residues written in upper case C. The first line consists of the leading symbol ‘>’ followed by the metpos ID. An arbitrary name can be entered after the symbol ‘#’ as separator. Files carry the names ‘methpos_metpos ID’. An example is given in Figure 2c. For bisulfite genomic sequencing data (11), methylation patterns in the above file format can be conveniently generated with the MethTools software suite (12).
Description of the web interface
MethDB can be accessed via WWW (http://www.methdb.de) (Fig. 3). For administrative reasons the home page is served by a commercial ISP. The database itself has its own dedicated web server. We recommend not to access this server directly since it can not be excluded that its IP number might change from time to time. The search page offers three forms for a structured search and one for a simple quick-search field.
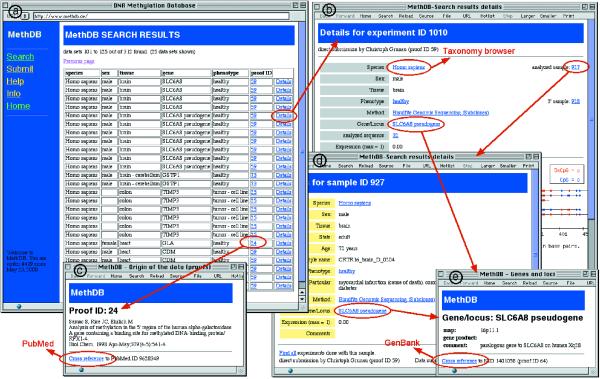
Figure 3.
Example for the output of MethDB after a search for methylation profiles and patterns. Initial result of a request to MethDB is a table (a). Each line of the table represents an individual experiment. A click on the ‘Details’ link brings the user to a window that shows detailed information about the experiment and displays the methylation data in a graphical form (b). Further clickable links provide information about, for instance, the reference for the data (c), the description of the investigated sample (d) or information about the gene that was studied (e). As examples for cross-references to external databases, the links to PubMed, GenBank and the Entrez Taxonomy browser are indicated.
Search for patterns and profiles. Methylation patterns (G,A,T,C,5mC sequences) and methylation profiles (level of methylation at cytosine sites across a sequence) are sequence specific data. It is possible to search the database for species, sex, tissue and a gene or locus. A table is returned with a list of matching records. The [Details]-link brings the user to a page with specific information about the particular experiment and the methylation profile or a list of methylation patterns. By definition, one experiment can deliver several patterns but only one profile. Multiple experiments can be performed on one sample. If an experimental method is DNA strand specific, the complement DNA strands are treated as separate samples.
Search for 5mC content. The query form permits to search for species, sex, tissue, phenotype and DNA type of a sample. The database returns a table of matching records together with their 5mC content data. The [Proof]-link brings the user to the reference for the data, the [Details]-link provides information about the sample and the experimental procedure used to obtain the data.
Search for phenotypes. The form allows to enter 5mC content values and to search for phenotypes that have been found to be associated with these 5mC content data. Provided enough background data is present this form might be useful for diagnostic purposes.
Quick search. In this simple query form a single search term can be entered. When the query is submitted a full text search is performed in the following fields: comment, method name, sample name, restriction enzyme, author, phenotype, tissue, gene and species name. In the resulting table, the most recent entries are listed first.
MethDB contains a number of cross references to other databases, for instance the NCBI Taxonomy browser (http://www.ncbi.nlm.nih.gov/Taxonomy/), Entrez PubMed, Entrez GenBank (http://www.ncbi.nlm.nih.gov/Entrez/) and OMIM (http://www.ncbi.nlm.nih.gov/Omim/)) and independent web sites. The tables ‘author’, ‘primer’ and ‘PCR’ are not yet accessible via the web-interface.
Submission policy and prospects of the database
Currently, the related literature is screened and the available data are entered into the database. Each time literature data has been entered, the corresponding authors of the original research papers are informed and asked to verify these entries. Data from unpublished sources can be submitted by personal communication to the database administrator. We would like to invite and encourage the scientific community to submit their findings. Naturally, for every database entry the author who has submitted the data is indicated. During the last year, the amount of knowledge about DNA methylation has increased considerably. However, while the mechanism of DNA methylation and the way it affects chromatin structure and gene expression becomes clearer and clearer the biological role of DNA methylation is far from being well-defined. The short-term goal of the MethDB project is to collect as much data as possible and to make these data available to the public. We believe that as soon as a sufficient amount of data has been accumulated it will be possible to gain new insights about the function of DNA methylation in the living cell. For this purpose it will be certainly necessary in the future to develop new software tools for the analysis of the data. We are willing to do this by ourselves or in a collaborative effort.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Susan Clark, Peter Warnecke, John Melki (CSIRO, Sydney), Igor Progribny (NCTR, Jefferson) and Melanie Ehrlich (Tulane University Medical Center, New Orleans) who provided unpublished data in an early stage of the development of MethDB. The work presented here was made possible by a grant of the Klaus Tschira Foundation (Heidelberg) to C.G. The authors are grateful to Niels Jahn and Maik Schilling for the introduction into database design.
References
- 1.Holliday R. (1989) DNA methylation and epigenetic mechanisms. Cell Biophys., 15, 15–20. [DOI] [PubMed] [Google Scholar]
- 2.Razin A. and Cedar,H. (1993) In Jost,J.P. and Saluz,H.P. (eds), DNA Methylation: Molecular Biology and Biological Significance. Birkhäuser Verlag, Basel, pp. 343–359.
- 3.Chen B., Kung,H.F. and Bates,R.R. (1976) Effects of methylation of the beta-galactosidase genome upon in vitro synthesis of beta-galactosidase. Chem. Biol. Interact., 14, 101–111. [DOI] [PubMed] [Google Scholar]
- 4.Razin A. (1998) CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bestor T.H. (1998) Gene silencing. Methylation meets acetylation. Nature, 393, 311–312. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki H., Allen,N.D. and Surani,M.A. (1993) In Jost,J.P. and Saluz,H.P. (eds), DNA: Molecular Biology and Biological Significance. Birkhäuser Verlag, Basel, pp. 469–477.
- 7.Goto T. and Monk,M. (1998) Regulation of X-chromosome inactivation in development in mice and humans. Microbiol. Mol. Biol. Rev., 62, 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz W.A. (1998) DNA methylation in urological malignancies Int. J. Oncol., 13, 151–167. [PubMed] [Google Scholar]
- 9.Maurer S.M., Firestone,R.B. and Scriver,C.R. (2000) Science’s neglected legacy. Nature, 405, 117–120. [DOI] [PubMed] [Google Scholar]
- 10.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5- methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunau C., Schattevoy,R., Mache,N. and Rosenthal,A. (2000) MethTools–a toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res., 28, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]