Abstract
Activation of innate antiviral responses in multicellular organisms relies on the recognition of structural differences between viral and cellular RNAs. Double-stranded (ds)RNA, produced during viral replication, is a well-known activator of antiviral defenses and triggers interferon production in vertebrates and RNAi in invertebrates and plants. Previous work in mammalian cells indicates that negative-strand RNA viruses do not appear to generate dsRNA, and that activation of innate immunity is triggered by the recognition of the uncapped 5′ ends of viral RNA. This finding raises the question whether antiviral RNAi, which is triggered by the presence of dsRNA in insects, represents an effective host-defense mechanism against negative-strand RNA viruses. Here, we show that the negative-strand RNA virus vesicular stomatitis virus (VSV) does not produce easily detectable amounts of dsRNA in Drosophila cells. Nevertheless, RNAi represents a potent response to VSV infection, as illustrated by the high susceptibility of RNAi-defective mutant flies to this virus. VSV-derived small RNAs produced in infected cells or flies uniformly cover the viral genome, and equally map the genome and antigenome RNAs, indicating that they derive from dsRNA. Our findings reveal that RNAi is not restricted to the defense against positive-strand or dsRNA viruses but can also be highly efficient against a negative-strand RNA virus. This result is of particular interest in view of the frequent transmission of medically relevant negative-strand RNA viruses to humans by insect vectors.
Keywords: deep sequencing, arbovirus, Dicer-2, AGO2, small RNA profiling
Viral infection is a serious threat for all organisms. As a result, host cells have evolved various strategies to sense and fight viral infections. A central molecular pattern betraying the presence of viruses in cells is double-stranded (ds)RNA, present in the viral genome, in viral replication complexes, or resulting from bidirectional transcription in DNA viruses. This molecular pattern triggers RNA interference in plants and invertebrates, and production of interferons (IFNs) in vertebrates (1). In the fruit fly, Drosophila melanogaster, antiviral RNAi relies on the recognition of dsRNA in infected cells by the RNaseIII enzyme Dicer-2 (Dcr-2). Dcr-2 processes the dsRNA into 21-bp siRNA duplexes. The siRNAs are transferred with the help of the dsRNA binding protein R2D2 to AGO2, a member of the Argonaute family. AGO2 is the central component of the RNAi silencing complex, which targets the viral RNA molecules for degradation (2). The importance of this pathway for the control of viral infections is illustrated by the strong susceptibility of Dcr-2, AGO2, and R2D2 mutant flies to infection by positive-strand and dsRNA viruses (3–6).
The negative-strand RNA viruses include some of the most important human pathogens, such as the hemorrhagic fever viruses Ebola and Lassa (up to 80% mortality), the Rabies virus (100% mortality), and the influenza virus. Apart from these viruses, which are transmitted between mammalian hosts, several negative-strand RNA viruses are transmitted to humans by insect vectors (arthropod-borne viruses or arboviruses). For example, Rift Valley Fever virus (RVF), a Bunyavirus that is transmitted by Aedes and Culex mosquitoes, can cause encephalitis and hemorrhagic fever (7). Another member of the Bunyavirus family, Crimean-Congo hemorrhagic fever virus, vectored by ticks and causing severe disease in humans (30% mortality), is endemic in many countries in Africa, Europe, and Asia. Therefore, it is of great interest to study the interaction of negative-strand RNA arboviruses with their insect host.
A systematic analysis of dsRNA production by different viruses in mammalian cells revealed that, whereas significant amount of dsRNA could be detected for viruses with dsRNA, positive-strand RNA or DNA genomes, this was not the case for the negative-strand RNA viruses tested (8). In these viruses, the nucleoprotein immediately associates with neosynthesized genomic RNA, thus preventing the formation of dsRNA (9). Analysis of the innate immune response against negative-strand RNA viruses in mammals revealed that triphosphate groups at the 5′ ends of the uncapped viral RNAs, rather than long dsRNAs, play a critical role in the induction of IFN synthesis (10–13). One may therefore wonder whether the Dcr-2–mediated antiviral response, which is triggered by dsRNA, can provide immunity against a negative-strand RNA virus in insect hosts. To address this question, we used the well-characterized insect model Drosophila and, as a prototype for negative-strand RNA arbovirus, the vesicular stomatitis virus (VSV), a member of the rhabdovirus family.
Here, we report that VSV does not produce readily detectable amounts of dsRNA in infected mammalian and Drosophila cells. Nevertheless, we observed a dramatic increase in the VSV titer in RNAi mutant flies, leading to the death of infected animals. Small RNA profiling of VSV infected flies identified viral siRNAs (vsiRNAs) equally matching both strands of the whole genome, consistent with their production from dsRNA. Our results extend the importance of RNAi as an antiviral host-defense mechanism in insects to the epidemiologically relevant group of negative-strand RNA viruses.
Results
VSV Does Not Produce Detectable Amounts of dsRNA in Mammalian and Drosophila Cells.
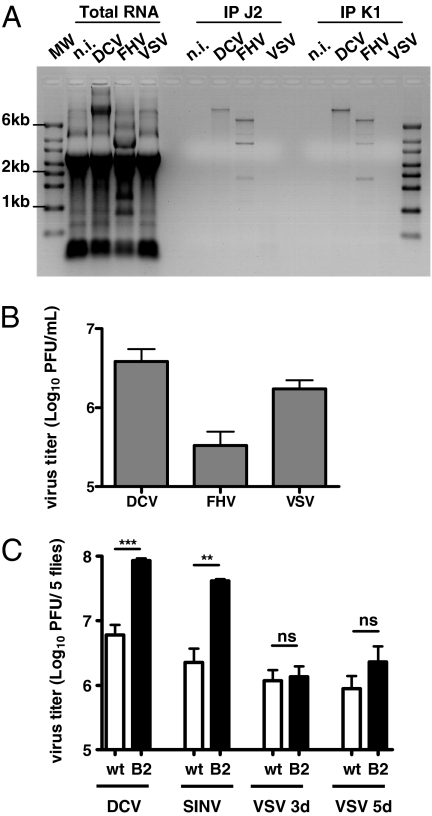
We first tested whether dsRNA can be detected in VSV-infected cells. Mammalian Vero cells were infected with VSV and immunostained with the anti-dsRNA monoclonal antibody J2. Double-stranded RNA was not detected, although cells were highly infected with VSV as visualized by immunodetection of the VSV-G protein (Fig. S1). We next analyzed the production of dsRNA in VSV-infected Drosophila cells. Immunostaining with the J2 antibody was not possible in the Drosophila cell lines tested because of a high background. As an alternative, total RNA from infected cells was isolated and dsRNA was immunoprecipitated using the J2 antibody or another dsRNA-specific monoclonal antibody, K1 (14). Drosophila Kc167 cells were infected with VSV or with the positive-strand RNA viruses flock house virus (FHV) and Drosophila C virus (DCV) as controls. The dsRNA precipitation revealed the presence of viral dsRNA in DCV- and FHV-infected cells (Fig. 1A). Similar results were obtained using the Drosophila S2 cell line (Fig. S2A). RT-PCR amplification of the immunoprecipitated RNAs and sequencing of the amplicons confirmed their viral origin (Fig. S2 B and C). In addition, the pulled-down RNAs were sensitive to digestion with the dsRNA-specific enzyme RNaseIII, but not to the single-stranded (ss) RNA specific RNasesA and T1 (Fig S2D). Double-stranded RNA was also readily detectable in cells infected with Cricket paralysis virus, a positive-strand RNA virus that encodes a suppressor of RNAi that does not interact with dsRNA (15) (Fig. S2E). In contrast, we did not detect dsRNA in VSV infected cells (Fig. 1A and Fig. S2A), even though the virus grew to similar titers as DCV or FHV (Fig. 1B and Fig. S2F). We further attempted to detect immunoprecipitated dsRNA in VSV-infected cells by RT-PCR, without any success, although we could detect the presence of both the genomic and the antigenomic strands in infected cells (Fig. S2G).
Fig. 1.
VSV does not produce detectable amounts of dsRNA in Drosophila despite efficient replication. (A) Immunoprecipitation with dsRNA-specific antibodies (J2, Center and K1, Right) of total RNA (Left) extracted from Kc167 cells infected with DCV [multiplicity of infection (MOI) 10], FHV (MOI 0.1), or VSV (MOI 10) at 4 d postinfection (dpi) for FHV and VSV and 6 dpi for DCV. Immunoprecipitates were subjected to agarose gel electrophoresis, which revealed dsRNA replication intermediates for DCV and FHV but not for VSV. MW, molecular weight marker; n.i., control. A representative experiment out of three is shown. (B) DCV, FHV, and VSV titers measured from supernatant of the infected Kc167 cells described in A before RNA extraction. The values represent the mean and SD of three independent experiments. (C) In contrast to DCV and SINV, VSV titer did not increase significantly in flies overexpressing B2 (hs-GAL4 > UAS-B2) compared with WT (hs-GAL4/+) flies. Flies were infected with 500 TCID50 DCV, 1,000 pfu SINV, and 5,000 pfu VSV. Viral titers were assayed from whole fly extract at 3 dpi (DCV, VSV) and 5 dpi (SINV, VSV). The values are the mean and SD of three independent groups of five flies. P values were calculated using Student's paired t test; **P < 0.01; ***P < 0.001; ns, not significant.
FHV expresses the viral suppressor of RNAi B2, which binds dsRNA with high affinity and prevents recognition and processing by Dcr-2 (16–18). Expression of B2 in transgenic flies decreases their resistance to DCV infection (6, 19). In addition, B2 overexpressing flies contained higher viral loads than control flies upon infection with DCV and Sindbis virus (SINV), supporting the evidence that dsRNA produced by those viruses trigger immunity. In contrast, overexpression of B2 did not affect the viral load of VSV at 3 d postinfection and only led to a mild and not significant increase in viral titer at 5 d postinfection (Fig. 1C). Taken together, our data suggest that VSV does not produce readily detectable amounts of dsRNA in flies, raising the question of the involvement of RNAi in the control of the infection.
RNAi Controls VSV Infection in Drosophila.
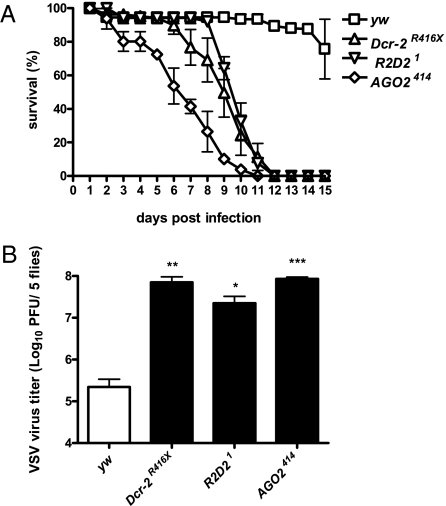
To address the potential role of RNAi in the control of VSV infection in vivo, we challenged wild-type (WT, yw and w1118) WT (yw and w1118), or Dcr-2 (Dcr-2R416X, Dcr-2L811Fsx), R2D2 (R2D21), and AGO2 (AGO2414) mutant flies with VSV. Whereas WT flies were resistant to VSV infection, mutant flies showed a severely compromised survival and died within 12 d (Fig. 2A and Fig. S3). The lethality was correlated with a 100- to 1,000-fold increase in viral titers, showing the inability of RNAi mutant flies to control viral replication (Fig. 2B). We conclude that RNAi plays a major role in the defense against VSV infection.
Fig. 2.
RNAi controls VSV infection in vivo. (A) RNAi mutants display a strong susceptibility to VSV challenge. Dcr-2R416X, R2D21, and AGO2414 mutant flies infected with 5,000 pfu VSV died within 12 dpi in contrast to the yw control flies. The values represent the mean and SD of at least three independent groups of 10 flies each. (B) The increased susceptibility was correlated with an increase in viral titer in all RNAi mutants compared with yw controls at the peak of viral replication (5 dpi). The values represent the mean and SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
RNAi Machinery Processes VSV dsRNA in Drosophila.
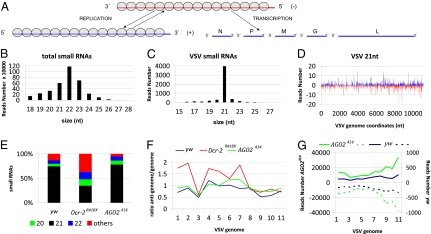
We next wondered whether VSV-derived siRNAs are produced during the course of infection, even though dsRNA could not be detected. We thus deep-sequenced small RNA libraries from WT and RNAi mutant flies. These experiments were performed independently in our two laboratories and will be referred to as Datasets 1 and 2 in Table S1. Flies were infected with 5,000 pfu VSV. Samples were collected at the peak of viral replication, 5 d postinfection (dpi) for Dataset 1 and 3 to 4 dpi for Dataset 2. Small RNA from 18 to 28 nt for Dataset 1, and from 19 to 24 nt from Dataset 2, were perfectly aligned against the Drosophila and the VSV genomes (Fig.3A). The total number of small RNA reads as well as VSV-derived reads are summarized in Table S1.
Fig. 3.
Virus-derived siRNAs in VSV infected flies. (A) Structure and organization of the VSV genome. The 11-kb long negative-strand RNA is the template for synthesis of five monocistronic mRNAs that encode the viral proteins N, P, M, G, and L. During replication, the negative-strand is the template for the synthesis of the full-length positive-strand RNA (antigenome), which will in turn serve as a template for the production of the genomic RNA. The N protein is tightly associated with the genome and antigenome RNA (gray circles). (B) Size distribution of total small RNAs in VSV-infected yw flies. (C) Size distribution of small RNAs aligned against the viral genome in VSV-infected yw flies. (D) Profile of 21 nt VSV-derived reads along the VSV genome. To comply with the orientation of the VSV reference sequence (NC_001560) used for all of the analyses, the genome is represented under its antigenomic polarity. Each VSV-derived small RNAs (vsiRNA) is represented by the position of its first nucleotide. The vsiRNAs matching the antigenome and the genome are shown in blue and red, respectively. The horizontal axis represents the antigenome coordinates. (E) Relative abundance of VSV-derived small RNAs of 20, 21, 22 nt and other sizes (19, 23–28 nt) in yw, Dcr-2R416X, and AGO2414 flies. (F) Relative strand representation of the 21 nt reads expressed as the ratio between the number of reads matching the antigenome over the number of reads matching the genome using 1,000 bp contiguous windows (1–11). (G) Representation of VSV-derived vsiRNAs from yw (blue) and AGO2414 (green) flies expressed as the sum of the reads over 1,000-bp contiguous windows covering the whole genome. The vsiRNAs matching the antigenome and genome are showed as solid and dotted lines, respectively. The sum of the reads corresponding to each window is represented in the vertical axis.
In WT flies (Dataset 1: yw and Dataset 2: w1118) (Table S1), the size distribution of total small RNA clearly shows enrichment at 20 to 23 nt (Fig. 3B and Fig. S4A). The pronounced peak at 22 nt is characteristic of miRNA that are processed by Dicer-1. In contrast, the VSV-derived small RNAs (vsiRNAs; 0.1% and 0.08% of total reads for Table S1, Datasets 1 and 2, respectively) are highly enriched in 21-nt long reads, which is the signature of Dcr-2 processing (Fig. 3 C and E and Fig. S4 B and D). The 21 nt vsiRNA matched equally along the entire genome and antigenome with a moderate enrichment at both the 5′ end of the genome and the 3′ end of the antigenome (Fig. 3 D and G and Fig. S4 C and F). The ratio of vsiRNAs derived from the genomic and antigenomic strands was close to 1 in WT flies (Fig. 3 D and F and Fig. S4 C and E). Because the VSV genome is 5 to 10 times more abundant than its antigenome in infected cells [(20); see also Fig. S2G], and because VSV mRNAs are produced in different amounts (9), these data suggest that the vsiRNAs are generated from dsRNA replication intermediates, rather than from internally transcribed mRNA or secondary structures.
Analysis of vs.iRNAs in Dcr-2 and AGO2 Mutant Flies.
Because Dcr-2−/− and AGO2−/− mutant flies display a strong susceptibility to VSV infection, we next investigated the involvement of these genes in the biogenesis and stability of VSV-derived vs.iRNAs. Small RNA libraries from VSV-infected Dcr-2−/− (Dataset 1: Dcr-2R416X; Dataset 2: Dcr-2L811fsx) and AGO2−/−(Table S1, Datasets 1 and 2: AGO2414) mutant flies were sequenced. Results are summarized in Table S1.
In AGO2−/− mutants, the number of VSV-derived vsiRNA reads was 100-fold higher than in wt flies (Figs. S5E and S6E). This finding correlates with the strong increase of viral RNA levels in AGO2−/− mutant flies as compared with WT (600-fold in the sample used for Dataset 1) (Table S1). Although small differences could be observed between the vsiRNAs sequenced in the two datasets, most likely reflecting the use of different 5′ and 3′ adapters to construct the libraries (21), the profiles were similar overall. We did not observe any strand bias in the vsiRNAs produced in AGO2−/− mutants and the antigenome-to-genome ratio was close to 1 (Fig. 3F for Dataset 1 and Fig. S4E for Dataset 2). The distribution and density along the viral genome and antigenome of AGO2−/− derived vsiRNAs were similar to those observed in infected WT flies, although a strong increase of vsiRNA reads mapping at the 5′ end of the genome (and the 3′ end of the antigenome) was observed (Fig. 3G and Fig. S4F). This phenomenon was already detectable in WT flies but was strongly amplified in AGO2−/− mutant flies, suggesting a preferential cleavage of viral dsRNA from the site of the initiation of replication.
Unexpectedly, VSV-derived small RNA peaks were identified in Dcr-2R416X and Dcr-2L811fsX mutant flies (Figs. S5B and S6B), indicating that some dicing (or random degradation) occurred even in the absence of functional Dcr-2. However, the number of VSV vsiRNA reads obtained in Dcr-2−/− flies was similar to that in WT flies, despite the dramatic increase in viral RNA in the mutant flies (350-fold in the sample used for Dataset 1) (Table S1). This finding indicates that antiviral dicing is strongly reduced in Dcr-2−/− mutant flies. In addition, the vsiRNAs profile of Dcr-2−/− flies was substantially altered compared with the profile obtained from WT or AGO2−/− mutant flies. In WT flies and in S2 cells (see below), at least 75% of vsiRNAs had a size of 21 bp; however, the size distribution of the vsiRNAs in Dcr-2−/− mutant flies was much more heterogeneous, with fewer than 40% of vsiRNAs having a size of 21 bp (Fig. 3E and Fig. S4D). Cluster analysis of the vsiRNA reads also revealed clear differences between the profile of Dcr-2−/− flies versus those of WT or AGO2−/− mutant flies and S2 cells (Fig. S7). In addition, although 21-nt reads were distributed along both strands of the viral RNA, the antigenome/genome ratio was biased toward the antigenome (overall ratio antigenome/genome = 1.2 in Dataset 1 and 2.9 in Dataset 2) (Table S1), and more pronounced at the 5′ end of the antigenome (Fig. 3F and Fig. S4E). This finding suggests that VSV-derived vsiRNAs in Dcr-2−/− flies are derived from mRNA rather than from dsRNA replication intermediates. Interestingly, whereas the profiles of small RNAs produced in WT and AGO2−/− mutant flies were similar overall in the two datasets, this was not the case for the Dcr-2−/− mutant flies, and the bias for siRNAs matching the 5′ end of the antigenome was much stronger in Dataset 2 than in Dataset 1 (Table S1). This observation may reflect the fact that two different Dcr-2 alleles were used, which may encode proteins with some residual activity.
VSV vsiRNAs Can Mediate Target Repression.
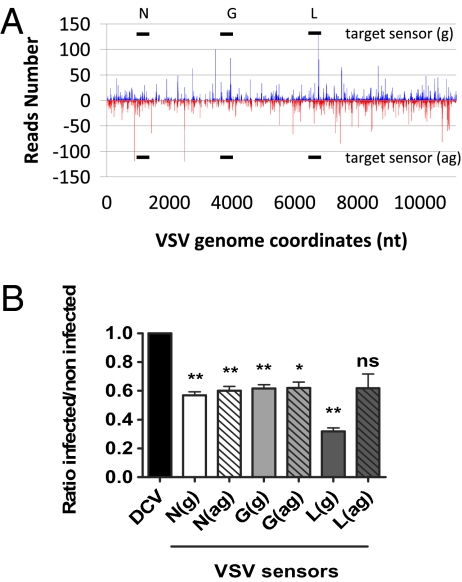
We next addressed the functional relevance of the VSV-derived vsiRNAs in the control of VSV infection in S2 cells. We first validated the RNAi-mediated control of VSV infection in this cell line by deep-sequencing small RNAs from S2 cells infected with VSV (MOI 10). The distribution and density of VSV-derived vsiRNAs along the viral genome was similar to those observed in WT infected flies (Fig. 4A and Fig. S8 A and B). To test the role of these vsiRNAs, we designed luciferase sensors containing 300-bp sense or antisense segments of several regions of VSV (Fig. 4A). These segments correspond to nt 736 to 1,027 (N), nt 3,478 to 3,783 (G), and nt 6,582 to 6,872 (L). As a control sensor, we used a 300-bp fragment of DCV (nt 6,207–6,507). The VSV and control sensor constructs were transfected into S2 cells and luciferase expression was monitored in the presence and absence of VSV infection.
Fig. 4.
VSV-derived vsiRNAs silence VSV sensors in S2 cells through RNAi. (A) Profile of 21 nt VSV-derived reads obtained from VSV-infected S2 cells as depicted in Fig. 3D. The three 300-nt regions of VSV selected in the N, G, and L genes to generate firefly luciferase sensors are shown in solid black lines. (B) Silencing of VSV reporters in VSV-infected cells. Each bar represents the ratio of luciferase expression measured between infected and noninfected cells normalized to the values obtained for the DCV control sensor (set at 1). The values represented are the mean and SD of three independent experiments. P values were calculated using Student's paired t test; *P < 0.05; **P < 0.01; ns, not significant.
Although VSV infection did not affect the luciferase expression levels in the DCV sensor, the VSV constructs were specifically silenced by at least 40% (Fig. 4B). Silencing was observed for both orientations of the VSV fragments, indicating that vsiRNAs can silence VSV by targeting both the genome and the antigenome. The strongest silencing was observed for the VSV-L sense construct (69%). The antisense L sensor construct was also silenced, but the effect was weaker and was not statistically significant in the format of our assay (Fig. 4B). To ensure that the observed silencing of the sensor construct was the result of RNAi, we investigated the dependence of the silencing on AGO2. We transfected AGO2 dsRNA in S2 cells to knock-down AGO2 expression, and observed a significant reversion of the silencing of the VSV-L sensor upon VSV infection (Fig. S8C). We conclude that the vsiRNAs produced in VSV infected cells can play an active role in the control of VSV RNA level.
Discussion
Most studies on antiviral RNAi in plants and invertebrates so far have focused on positive-strand RNA viruses (3, 5, 6, 22–31). We show here that RNAi is also a major antiviral response in vivo against the negative-strand RNA virus VSV in the model insect Drosophila. Indeed, flies mutant for the genes encoding the core components of the RNAi pathway Dcr-2, R2D2, and AGO2 are no longer able to control VSV replication and display a more than two-log increase in viral titer compared to WT. The recently described component of the RNAi pathway Ars2 is also required to control VSV replication in Drosophila cells (32). These data are in line with previous studies in the nematode Caenorhabditis elegans, which reported a 4-fold to 10-fold increase of viral titer in VSV-infected embryonic primary cell cultures derived from RNAi mutants compared with WT (33, 34). We further show that as a consequence of the uncontrolled viral replication, survival of RNAi mutant flies is severely compromised. Taken together, these data indicate that RNAi in flies controls infection by a negative-strand RNA virus and, more generally, represents a major host-defense pathway against RNA viruses, irrespective of their genome. Whether RNAi also participates in the control of DNA virus infections in invertebrates, as previously reported in plants, remains to be investigated (35, 36).
A key question in innate immunity is that of the microbial molecule or pattern triggering the host-response. In the case of RNAi, Dicer enzymes recognize dsRNA either as extended secondary structures on viral RNA molecules, or dsRNA generated during the course of viral replication (i.e., replicative intermediates). In plants, where studies have focused on positive ssRNA viruses, there is some evidence for the first scenario. For example, infection by the Tombusvirus CymRSV generates ∼80% to 90% of viral small RNAs of sense polarity, which may reflect the excess of genome versus antigenome RNA present in infected cells (24, 27, 30). However, the profiles of vsiRNAs produced in response to several other plant viruses are consistent with processing of a dsRNA precursor: these profiles are characterized by an approximately equal number of vsiRNAs matching the positive or the negative strand of the viral RNA, and covering the whole length of the viral genome (24, 26, 29). In invertebrates, small RNA profiling in cells or animals (Drosophila, Aedes mosquitoes, and C. elegans) infected with positive ssRNA or dsRNA viruses supports the idea that vs.iRNAs are generated from viral dsRNA intermediates of replication (22, 23, 28, 31, 37). In particular, vsiRNAs generated in cells infected by SINV, Drosophila A virus (DAV), Drosophila X virus (DXV), Drosophila totivirus (DTV), or Drosophila birnavirus (DBV) evenly cover both strands of the whole genome. In contrast, in the case of FHV, the number of reads mapping to the 5′ and 3′ ends of the viral RNAs are increased, suggesting that the extremities of the genome, where replication starts, are preferentially diced (22). Interestingly, the ends of negative-strand RNA virus genome and antigenome RNA can base pair to form a panhandle structure (38), which plays a critical role in induction of IFN in mammalian cells, and may trigger RNAi (39–41). However, the profiles obtained in VSV-infected WT cells or flies do not support this hypothesis, and we did not observe any increase in the number of vsiRNAs matching the ends of the VSV RNAs. Overall, the profiles we observed are remarkably similar to that of SINV, DAV, DXV, DTV, or DBV. This result leads us to propose that the VSV vsiRNAs are generated from dsRNA, rather than from secondary structures on genome or messenger RNAs, and that the VSV RNA features recognized by the mammalian and insect innate immune systems are different. This conclusion is based on data from the highly sensitive technique of deep-sequencing of small RNAs, and we infer that our inability to detect dsRNA using standard methods (8) reflects the fact that the amount of dsRNA produced in VSV-infected cells is below either the detection limit of the antibodies used or the FHV B2 protein binding capacity. An alternative yet intriguing possibility is that Dcr-2, of which little is known in the cellular context, could displace the viral nucleoprotein to gain access to dsRNA. Such a function may involve the helicase activity of Dcr-2, the role of which in RNAi remains poorly understood (42).
Of central interest to us in this study was the comparative analysis of the small RNA profiles in WT and RNAi mutant flies, a question which to the best of our knowledge has not been addressed earlier. The profile of vsiRNAs observed in AGO2−/− mutant flies strongly resembles the one obtained in WT infected flies, although the number of reads is 100-fold higher. Despite this intense dicing activity, AGO2−/− mutant flies are as susceptible to VSV infection as Dcr-2−/− mutant flies, indicating that dicing viral dsRNA per se does not represent an efficient defense against viral infections. The similarities observed in WT and AGO2−/− profiles strongly suggest that the absence of AGO2 does not affect Dcr-2 activity, precluding a coordinate action of both antiviral players.
Our analysis also revealed unexpected VSV-derived small RNAs in the Dcr-2−/− mutant flies, although their number was strongly reduced compared with WT after normalization by viral titers in both conditions. These small RNAs are enriched in the sizes 19 to 23 nt, and therefore are not likely to represent solely random degradation products of viral RNAs. Importantly, the two Dcr-2 alleles used contain premature stop codons upstream of the RNaseIII domains, and are considered as genetic nulls. In addition, the number, size distribution and coverage along the VSV genome of these small RNAs clearly differ from those observed in WT or AGO2−/− mutant flies. More specifically, we observed a strong bias of small RNAs towards the positive strand in Dcr-2−/− mutant flies and a 5′ to 3′ gradient of small RNAs along the viral antigenome. These data correlate with the relative amount of viral transcripts and suggest that in the absence of Dcr-2, small vsiRNAs are generated from viral mRNAs rather than from viral intermediates of replication. Furthermore, we also observed an enrichment of reads matching the extremities of both the genome and the antigenome, which contain short structured regions involved in the regulation of viral RNA synthesis (43). Residual Dcr-2 activity may contribute to the generation of small RNAs in mutant flies, as suggested by the differences observed between the profiles obtained for the two Dcr-2 alleles used. Taken together, our data indicate that Dcr-2−/− mutant flies still produce VSV-derived small RNAs. These small RNAs do not appear to play a significant role in the control of the antiviral response, as illustrated by the spectacular increase in viral titer observed in Dcr-2−/− flies. A possibility that warrants further investigation is that the product of the second Drosophila Dcr gene, the miRNA-producing enzyme Dcr-1, processes viral RNA secondary structures to generate these small RNAs. The presence of Dcr-2-independent small RNAs is reminiscent of the situation in virus-infected Arabidopsis, where virus-derived small RNAs are produced by Dicer-like (DCL)1 and DCL3 in the absence of the antiviral DCL2 and DCL4. As we observed with flies, these DCL2/-4 independent small RNAs do not contribute significantly to antiviral defense (26, 44).
In conclusion, we have shown that RNAi can control the negative-strand RNA arbovirus VSV, indicating that dsRNA produced in cells infected with a negative-ssRNA virus can trigger antiviral immunity. These findings were mostly inferred from virus-derived small RNA profiles extracted from our deep sequencing data, stressing the relevance of high-throughput sequencing technologies to characterize antiviral responses and host pathogen interactions. Overall, the profiles of vsiRNAs from Drosophila cells infected by positive ss, ds, or negative ss RNA viruses are remarkably similar, pointing to a common mechanism of processing, despite the sharp differences in strategies used by these viruses to replicate.
Materials and Methods
Infection of Flies and Cells.
Information regarding the mutant strains used, the tissue culture conditions, and propagation and titration of viral stocks can be found in the SI Materials and Methods. Infection by intrathoracic injection was performed as described previously (3).
RNA Isolation and Immunoprecipitation of dsRNA.
For dsRNA detection, 1.5 × 107 S2 or Kc167 cells were infected with VSV (MOI 10), DCV (MOI 10), or FHV (MOI 0.1). Cells were collected in 15-mL reaction tubes and total RNA was extracted using TriReagent (Gibco-BRL) according to the manufacturer's instructions. RNA was quantified and immunoprecipitation was performed as previously described (45). In brief, 40 μg of total RNA was incubated overnight at 4 °C in polysomal lysis buffer with 10 μg J2 or K1 antibody (Scicons). Then, 50 μL of protein A-agarose solution (Invitrogen) was added, and incubation continued at 4 °C for 4 h. Complexes were washed eight times in polysomal lysis buffer and, after degradation of the protein complexes by proteinase K digestion (30 min at 50 °C), RNA was recovered by phenol-chloroform extraction and ethanol precipitation. The RNA pellet was resuspended in 10 μL RNase-free water.
Sequencing, Assembly, and Analysis of Small RNA Libraries.
The small RNA library of S2 cells and whole flies were constructed as described (46) and sequenced by the Illumina Genome Analyzer II. Reads were then aligned to a reference consisting of the VSV genome from National Center for Biotechnology Information (NCBI) (accession number NC_001560) using the Bowtie program with standard parameters in genome assembly. Reads aligning to the VSV genome with zero mismatches were retained and analyzed using in-house Perl scripts and Excel. Sequences were submitted to the NCBI Small Read Archive under the accession number SRP002753.
Silencing of a VSV Sensor RNA.
Construction of the sensor plasmids and monitoring of their activities were done using standard protocols as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratories for helpful discussions and technical advice. We thank Odile Sismeiro, Jean-Yves Coppee (Plateforme Transcriptome et Epigénome, Institut Pasteur), Stéphanie Le Gras, and Bernard Jost (Plateforme Biopuces et Séquençage, Institut de Génétique et de Biologie Moléculaire et Cellulaire) for technical support with deep sequencing, and Monique Lafon for helpful advice and discussion on negative-strand RNA viruses. We thank R. Carthew (Northwestern University, Evanston, IL) and M. Siomi (Keio University, Tokyo) for providing the Dcr-2 and AGO2 fly stocks, respectively. This work was financially supported by a Centre National de la Recherche Scientifique Action Thématique et Incitative sur Programme starting grant (to S.P.), Agence Nationale de la Recherche Grants ANR-09-JCJC-0045-01 (to M.-C.S.) and ANR-09-MIEN-006-01 (to J.-L.I.), European Research Council Grant ERC 242703 (to M.-C.S.), the Balzan Foundation (to J.A.H.), and National Institutes of Health Grant PO1 AI070167 (to J.-L.I. and J.A.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences were submitted to the National Center for Biotechnology Information Small Read Archive under the accession no. SRP002753.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014378107/-/DCSupplemental.
References
- 1.Kemp C, Imler JL. Antiviral immunity in Drosophila. Curr Opin Immunol. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 4.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouloy M, Weber F. Molecular biology of Rift Valley fever virus. Open Virol J. 2010;4:8–14. doi: 10.2174/1874357901004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conzelmann KK. Nonsegmented negative-strand RNA viruses: Genetics and manipulation of viral genomes. Annu Rev Genet. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 10.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 11.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 12.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 13.Plumet S, et al. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schönborn J, et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nayak A, et al. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 17.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 18.Fagegaltier D, et al. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci USA. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS ONE. 2009;4:e5866. doi: 10.1371/journal.pone.0005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whelan SP, Wertz GW. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: Identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linsen SE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 22.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donaire L, et al. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 2009;392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Flynt A, Liu N, Martin R, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci USA. 2009;106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Ruiz H, et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molnár A, et al. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang XB, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szittya G, et al. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010;6:e1000838. doi: 10.1371/journal.ppat.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Q, et al. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci USA. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabin LR, et al. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 35.Blevins T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Parameswaran P, et al. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grinnell BW, Wagner RR. Nucleotide sequence and secondary structure of VSV leader RNA and homologous DNA involved in inhibition of DNA-dependent transcription. Cell. 1984;36:533–543. doi: 10.1016/0092-8674(84)90246-0. [DOI] [PubMed] [Google Scholar]
- 44.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 45.Peritz T, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 46.Pfeffer S. Identification of virally encoded microRNAs. Methods Enzymol. 2007;427:51–63. doi: 10.1016/S0076-6879(07)27003-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.