Abstract
Predictions of microRNA-mRNA interactions typically rely on bioinformatic algorithms, but these algorithms only suggest the possibility of microRNA binding and may miss important interactions as well as falsely predict others. We developed an affinity purification approach to empirically identify microRNAs associated with the 3′UTR of the mRNA encoding Hand2, a transcription factor essential for cardiac development. In addition to miR-1, a known regulator of Hand2 expression, we determined that the Hand2 3′UTR also associated with miR-133a, a microRNA cotranscribed with miR-1 in cardiac and muscle cells. Using a sequential binding assay, we showed that miR-1 and miR-133a could occupy the Hand2 3′UTR concurrently. miR-133a inhibited Hand2 expression in tissue culture models, and miR-133a double knockout mice had elevated levels of Hand2 mRNA and protein. We conclude that Hand2 is regulated by miR-133a in addition to miR-1. The affinity purification assay should be generally applicable for identifying other microRNA-mRNA interactions.
Keywords: heart, cardiomyocytes, C2C12, MS2
MicroRNAs have assumed an increasingly recognized importance in the control of mammalian gene expression. Nowhere has this been more evident than in the cardiovascular system, where these molecules have been shown to contribute critically to normal heart development and responses to pathogenic stimuli (for review, see ref. 1). Targeted deletion of Dicer leads to severe cardiac dysfunction (2), and microRNA pathways have been proposed as possible therapeutic targets (3). One of the best examples of microRNA regulation in the heart involves the basic helix-loop-helix transcription factor, Hand2, and its regulation by miR-1 (4). As in other systems, however, microRNA networks in the heart are quite complex, largely because individual microRNAs can have hundreds of mRNA targets and each target can, potentially, be regulated by dozens of microRNAs. In plants, microRNAs and their recognition elements (termed MREs) have extensive complementarity, making microRNA targets relatively easy to identify. In contrast, the incomplete complementarity between microRNAs and their MREs in mammalian systems introduces considerable uncertainty to the association of particular microRNAs and their mRNA targets (5).
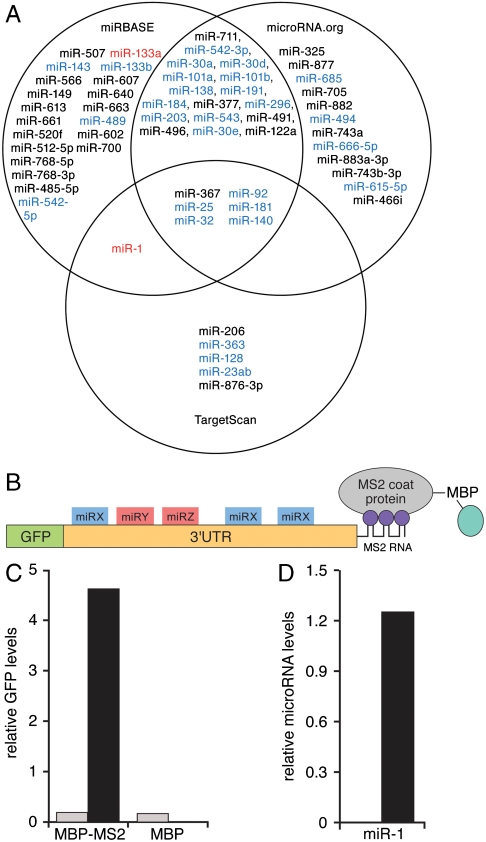
The original characterization of microRNA signaling in Caenorhabditis elegans posited that the prototype microRNA, lin-4, bound to multiple sites within the 3′UTR of its mRNA target, lin-14 (6, 7). By analogy, it is highly unlikely, given the properties of microRNAs and the fundamental importance of Hand2 in cardiac development, that the single MRE previously identified in the Hand2 3′UTR as a binding site for miR-1 is the only such element involved in regulating Hand2 expression. One possibility is that the Hand2 3′UTR contains additional miR-1 MREs that have not been identified (in a manner similar to the reiterated lin-4 binding sites in lin-14). Alternatively, Hand2 might be controlled through other microRNAs, each with their own binding sites. An examination of three prediction algorithms suggested that, in addition to miR-1, the 816 nucleotide long Hand2 3′ UTR could potentially interact with up to 60 other microRNAs (Fig. 1A). There was little overlap among these algorithms, however, and which of these predicted sites actually bind to a microRNA in vivo is unknown.
Fig. 1.
Characterization of Hand2-associated microRNAs. (A) Venn diagram of predicted Hand2 3′UTR-associated microRNAs from three algorithms [miRBase (now MicroCosm), TargetScan, and microRNA.org]. MicroRNAs expressed in heart are shown in blue. miR-1 and miR-133a, identified in the affinity purification assay (see below) are shown in red. MicroRNAs predicted to regulate Hand2 but not expressed in heart are shown in black. (B) Schematic of the MS2 affinity purification assay. An MS2-tagged Hand2 3′UTR is expressed downstream of a GFP reporter. Purification of the transcript is mediated by the affinity of the MS2 coat protein (expressed as a fusion with maltose binding protein, MBP) for the MS2 RNA sequence tag. (C) Real-time PCR quantification of GFP-Hand2 3′UTR reporter transcripts expressed in HEK293 cells and purified on an MBP-MS2 affinity column. MBP represents a column lacking the MS2 binding protein. MS2-tagged and untagged transcripts are shown in black and gray, respectively. (D) Taqman PCR analysis of miR-1 associated with the Hand2 3′UTR expressed in differentiated C2C12 cells and purified on an MBP-MS2 affinity column. No binding was detected when the Hand2 3′UTR was inserted in reverse orientation.
An underlying problem in predicting microRNA interactions is that they depend upon relatively short stretches of complementarity (5). The “seed” sequence, located at the 5′ end of the microRNA, is the primary determinant of binding, but additional elements of homology at the 3′ end of the microRNA can compensate for mismatches within the seed region (8, 9) and bioinformatic algorithms are continually evolving to incorporate such nuances of microRNA recognition. Importantly, the algorithms designed to predict microRNA binding do not take into account whether an mRNA and microRNA are coexpressed within the same cellular compartment. Additionally, proteins have been identified that can bind to certain MRE sequences and block microRNA interactions in a cell-specific or signal-responsive manner (10). Thus, microRNAs that interact with a particular MRE in one cellular state may not interact in another. MicroRNA interactions that occur in the absence of seed sites have also been described (11). Taken together, these observations suggest that the mere presence of a seed site may not be sufficient or necessary for microRNA regulation (8, 11–13) and point out the need for empirical approaches for identifying microRNA interactions.
The transcription factor, Hand2, provides a particularly good model for studies of microRNA regulation. In the early stages of cardiac development, Hand2 is required for proliferation of ventricular cardiomyocytes. Hand2 expression must be shut off for further stages of differentiation, however, and this step is controlled by miR-1 (4). Overexpression of miR-1 in the hearts of transgenic mice causes a ventricular defect that has been attributed to premature differentiation and early withdrawal of cardiomyocytes from the cell cycle. As a result, these mice have thin-walled hearts and develop heart failure. Genetic deletion of miR-1 causes the opposite phenotype, that is, hyperplasia and thickened chamber walls, as well as ventricular septal defects (VSDs) (14).
The miR-1 precursor generates another microRNA, miR-133a (15, 16), whose genetic deletion in heart also causes neonatal death with large VSDs, increased apoptosis and fibrosis, and dilated right ventricles (17). Because loss of miR-1 and miR-133a cause similar phenotypes, it would be reasonable to predict that the two microRNAs might, at least in some instances, also share some common targets. However, no common mRNA targets of these miRNAs have been identified and, among the many algorithms developed to predict microRNA targets [miRBase (now called MicroCosm), Targetscan, Pictar, microrna.org, etc.), only miRBase predicts that both miR-1 and miR-133a interact with the Hand2 3′UTR.
To address whether Hand2 might be regulated by additional microRNAs, we developed an affinity purification approach for identifying mRNA-associated microRNAs empirically. Using this method, we showed that the Hand2 3′UTR associated with multiple microRNAs and that one of the most robust interactions involved miR-133a. Furthermore, we showed that the binding of miR-133a and miR-1 to the Hand2 3′UTR could occur concurrently to mediate combinatorial control. Finally, we demonstrated that miR-133a regulates Hand2 expression in both tissue culture models and in hearts of miR-133a double knockout mice.
Results
Affinity Purification of Hand2 3′UTR Complexes.
Hand2 is one of the best-characterized microRNA targets in the heart. Because some mRNA targets are controlled by multiple microRNAs, we sought to determine whether miR-1 was the only microRNA responsible for regulating Hand2 expression. To assess which of the candidate MREs in the Hand2 3′UTR have the potential to contribute to regulation, we first assayed microRNA levels in primary rat neonatal cardiomyocytes using the Applied Biosystems (ABI) Taqman multiplex microRNA array. This array detected more than 200 microRNAs, the most abundant of which are plotted in Fig. S1. Of note, most of the microRNAs corresponding to predicted Hand2 MREs were not expressed at appreciable levels in primary cardiomyocytes, so these (shown in black in Fig. 1A) were eliminated from further consideration.
To determine which of the microRNAs expressed in heart had the potential to regulate Hand2 expression, we utilized an affinity purification method. Although microRNAs can bind to 5′ UTR sequences and coding regions of some mRNAs (18–20), we chose to focus on the Hand2 3′UTR, as this region is most commonly involved in mediating microRNA function. This region was fused to an MS2 binding site and cloned downstream from a GFP reporter (Fig. 1B). We generated an affinity column by expressing the bacteriophage MS2 binding protein as a fusion with maltose-binding protein (MBP), which was then bound to amylose beads. The MS2-tagged Hand2 3′UTR or control Hand2 sequences lacking the MS2 tag were expressed in HEK293 cells, transcripts were isolated in a nondenaturing lysis buffer, and the extracts were applied to the MBP-MS2 column or a column containing MBP alone as a negative control. Transcripts were purified from these columns using Trizol and were identified by RT-PCR mediated detection of the GFP reporter. The MS2-tagged reporter transcripts were purified only from MBP-MS2 columns and not from columns containing the MBP component alone (Fig. 1C). In addition, Hand2 reporter constructs lacking the MS2 tag did not associate with either the MBP-MS2 or MBP columns, confirming the specificity of the affinity purification assay.
To determine whether this method could detect endogenous microRNAs associated with the MS2-tagged Hand2 3′UTR, we tested the system in C2C12 cells. These cells are mesenchymal precursors that do not express muscle proteins or the muscle-specific microRNAs, miR-1 and miR-133a, in the presence of serum. When serum is removed from the culture medium, these cells differentiate into muscle and express both microRNAs (15). C2C12 cells were transiently transfected with the MS2-tagged GFP-Hand2 3′UTR, differentiated by serum starvation, lysed, and mRNA-miRNA complexes were purified over an MBP-MS2 affinity column. After determining by RT-PCR that the GFP reporter was purified on this column, we assayed for associated miR-1 using the Taqman RT-PCR method. The results confirmed that miR-1 associated with the Hand2 3′UTR (Fig. 1D). Importantly, miR-1 did not copurify with an MS2-tagged 3′UTR in reverse orientation, demonstrating the specificity of the MS2 purification strategy for identifying target-specific microRNA interactions.
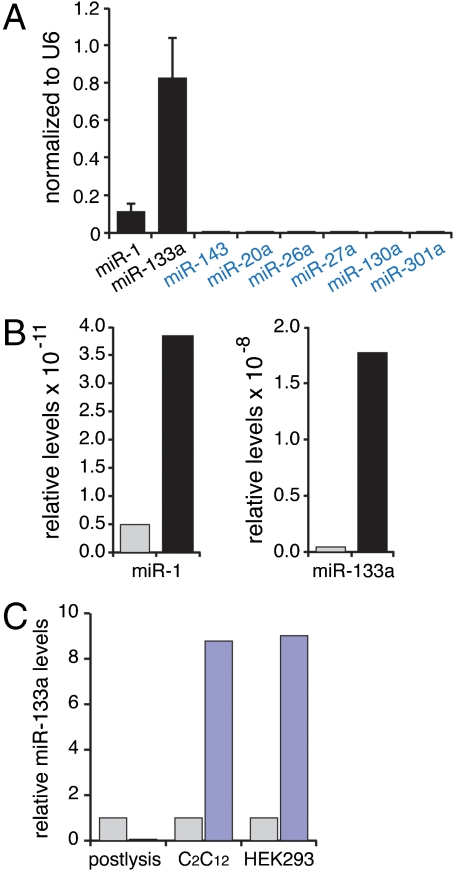
Although it would be possible to test the associations of each of the microRNAs depicted in Fig. 1A individually using the protocol described above, this approach is laborious and could miss interactions that were not predicted by any of the algorithms. For the MS2 affinity purification assay to be useful as a screen for unsuspected microRNA interactions, a nonbiased method would be preferable. We consequently modified the binding assay to incorporate a multiplex PCR analysis of potentially interacting microRNAs. Dissociated rat neonatal cardiomyocytes were infected with a lentivirus expressing the MS2-tagged Hand2 3′UTR reporter, and complexes were purified as described above. To identify associated microRNAs, we used the ABI Taqman multiplex microRNA array, which allows analysis of several hundred microRNAs simultaneously from a single RT-PCR reaction. One of the identified microRNAs was miR-1, confirming the fidelity of our assay (Fig. 2A). A considerably stronger interaction, however, was observed for miR-133a, and this was confirmed when the binding assays were repeated using specific Taqman primers (Fig. 2B). Notably, several abundant microRNAs were not detected in the affinity purified complexes, including miR-143 (predicted to interact by miRBase, but not microRNA.org, PicTar, or TargetScan; Fig. 1A). This result is consistent with the findings of others that the mere coexistence of a predicted MRE and cognate microRNA does not necessarily ensure that the two components will associate (12, 13).
Fig. 2.
Identification of Hand2-associated microRNAs by multiplex Taqman PCR. (A) miR-1 and miR-133a association was identified by multiplex PCR analysis of primary cardiomyocytes infected with lentiviruses expressing the GFP-Hand2 3′UTR-MS2 reporter. Six microRNAs, shown in blue, were abundantly expressed in heart, but did not associate with the Hand2 3′UTR probe. Results represent four independent experiments. Data are normalized to a U6 internal control represented on the array. (Mean ± SE are shown.) (B) Individual Taqman PCR confirmations of microRNA binding to probes containing the Hand2 3′UTR or reverse 3′UTR in primary cardiomyocytes. Hand2 3′UTR in correct and reverse orientation are shown in black and gray, respectively. Note scale differences in the two graphs. Results are representative of four independent experiments. (C) miR-133a interaction does not occur postlysis. Extracts from HEK293 cells expressing the MS2-tagged Hand2 3′UTR were mixed 1∶1 with extracts from differentiated C2C12 cells. No miR-133a binding was detected in the mixed extracts (left lanes). Differentiated C2C12 cells transfected with the MS2-tagged Hand2 3′UTR (middle lanes) or HEK293 cells expressing the MS2-tagged Hand2 3′UTR and transfected with miR-133a (right lanes) showed binding of miR133a in a 3′UTR-dependent manner. Blue and gray bars represent transcripts containing or lacking the Hand2 3′UTR, respectively. Similar results were obtained in three separate experiments.
To rule out the possibility that miR-133a and Hand2 associate after cell lysis, we expressed the MS2-tagged Hand2 3′UTR in HEK293 cells (which do not normally express miR-133a) and mixed lysates from these cells with lysates from miR-133a expressing differentiated C2C12 cells that did not contain the Hand2 probe (Fig. 2C). The MS2-tagged Hand2 mRNA was then affinity purified from the mixture and assayed for miR-133a binding. No association of miR-133a to the Hand2 3′UTR was detected in the postlysis mixtures. In contrast, when the Hand2 probe was introduced into differentiated C2C12 cells or HEK293 cells transfected with miR-133a, binding was readily apparent. These studies indicate that the association between the Hand2 3′UTR and miR-133a requires that the two are expressed in the same cell and indicates that binding does not occur after lysis.
Characterization of miR-133a Binding to the Hand2 3′UTR.
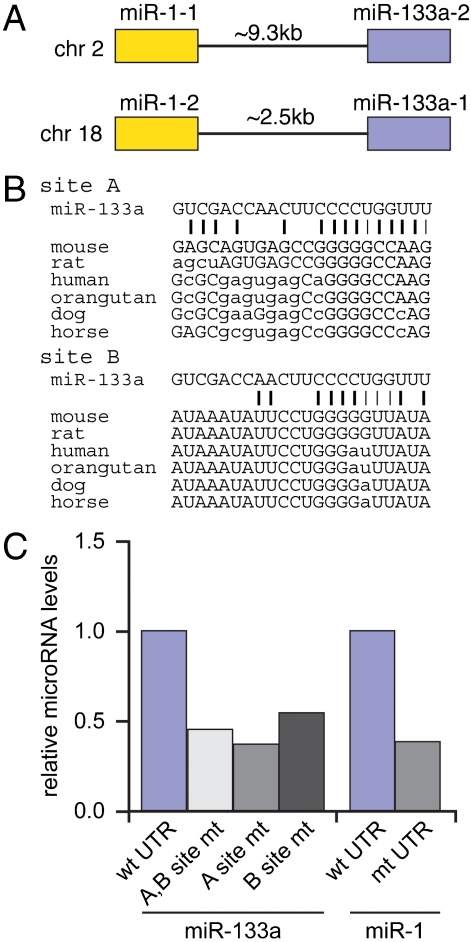
In mice, miR-1 and miR-133a are produced in the heart from two bicistronic precursors, one on chromosome 2 that expresses miR-1-1 and miR-133a-2 and the other on chromosome 18 that expresses miR-1-2 and miR-133a-1 (Fig. 3A). Although deletion of miR-133a-1 and miR-133a-2 individually in mice has no effect, deleting both together causes a cardiac hyperproliferation phenotype that is reminiscent in some respects to that of the miR-1-2 knockout (17).
Fig. 3.
Identification of miR-133a binding sites. (A) miR-1 and miR-133a are coexpressed from two separate genomic loci. (B) Alignment of miR-133a with sequences from the Hand2 3′UTR reveals two potential seed sites. Site A was identified by miRBase but not by three other microRNA prediction algorithms. Site B was identified by visual inspection of the Hand2 3′UTR. (C) Mutation of the A and B sites within the full-length Hand2 3′UTR decreases miR-133a binding to the Hand2 3′UTR in primary cardiomyocytes. Binding assays were also performed using a miR-1 MRE mutant. Results are representative of three separate experiments.
As mentioned above, only miRBase identified a potential miR-133a MRE within the Hand2 3′UTR (Fig. 1A). Neither this sequence (site A in Fig. 3B) nor a second potential site (site B) were sufficiently homologous to miR-133a to allow identification by three other prediction programs, however. To test whether the two putative miR-133a MREs were indeed responsible for the miR-133a interaction, we mutated these elements in the context of a GFP-Hand2 3′UTR lentivirus and monitored binding in primary rat cardiomyocytes. Binding assays employing a mutated miR-1 MRE (4) were also performed for comparison. Mutating the putative miR-133a elements, individually or in combination, substantially decreased the association of endogenous miR-133a, as compared to a virus containing a wild-type Hand2 3′UTR (Fig. 3C). Thus, it is likely that both elements contribute to miR-133a binding. Because the binding was not completely abolished upon mutation of the two MREs, it is possible that additional elements also capable of mediating the interaction with miR-133a remain to be identified. The loss of binding that resulted from the mutation of the miR-133a sites was equivalent to that seen after mutating the previously characterized miR-1 site.
Sequential Purification of miR-1 and miR-133a with the Hand2 3′UTR.
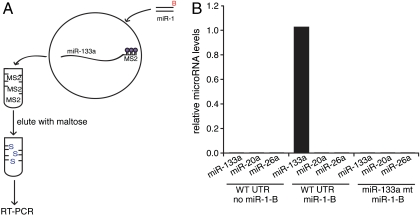
The idea that miR-1 and miR-133a might cooperate to regulate Hand2 expression implies that they interact with the same 3′UTR simultaneously. Our affinity purification assay provides a means to directly test this possibility. Primary cardiomyocytes were transfected with a biotinylated miR-1 oligonucleotide and simultaneously infected with viruses expressing MS2-tagged versions of either the wild-type Hand2 3′UTR or a mutant lacking miR-133a binding sites (Fig. 4A). Hand2 3′UTR complexes were purified on an MBP-MS2 affinity column to eliminate unbound microRNAs and the biotinylated miR-1 associated with other transcripts. Eluates from this column were then applied to a streptavidin column to isolate the MS2-tagged transcripts that also associated with biotinylated miR-1. Complexes from the second column, containing both biotinylated miR-1 and the Hand2 3′UTR, were then eluted and assayed for miR-133a using the Taqman RT-PCR method. miR-133a was detected in complexes associated with biotinylated miR-1, but only if the miR-133a MREs were intact (Fig. 4B), indicating that miR-133a and miR-1 must be capable of binding to the same Hand2 3′UTR transcript simultaneously. As a control, two other microRNAs (miR-20a and miR-26a) present in cardiomyocytes but not predicted to bind to the Hand2 3′UTR were also tested. No binding was detected for either, even in the presence of the biotinylated miR-1. Reporter assays performed in HEK293 cells demonstrate that this simultaneous occupancy facilitates synergistic regulation by miR-1 and miR-133a (Fig. S2).
Fig. 4.
miR-1 and miR-133a associate with the Hand2 3′UTR simultaneously in primary cardiomyocytes. (A) Schematic diagram of the sequential pulldown of biotinylated miR-1 and miR-133a with the Hand2 3′UTR. (B, Left) In the absence of biotinylated miR-1 (no miR-1-B), no microRNAs are purified in the presence of the wild-type Hand2 3′UTR (WT UTR). (Middle) In the presence of miR-1-B and WT UTR, miR-133a is enriched, but not miR-20a or miR-26a (two cardiac microRNAs not predicted to bind to the Hand2 3′UTR). (Right) Sequential pulldown using the miR-133a double MRE mutant did not copurify miR-133a. Results represent two independent experiments.
miR-133a Inhibits Hand2 Expression.
To determine whether miR-133a regulates Hand2 expression, we first performed reporter assays in nondifferentiated C2C12 cells. Introduction of double-stranded miR-133a oliogonucleotide mimics markedly reduced expression of reporter constructs that included the Hand2 3′UTR (Fig. 5A). No decrease was detected in reporter constructs lacking the 3′UTR. Conversely, antisense oligonucleotides directed against miR-133a relieved repression of a luciferase reporter in primary cardiomyocytes (Fig. S3). We next asked whether the endogenous Hand2 in cardiomyocytes was regulated similarly. Primary cardiomyocytes were transfected with miR-133a-blocking oligonucleotides, lysed, and Hand2 protein and mRNA levels examined by Western blotting and RT-PCR (Fig. 5 B and C). The blocking oligonucleotides increased levels of both Hand2 protein and mRNA. This result differs from that reported for miR-1, which appears to regulate Hand2 only at the protein level (14).
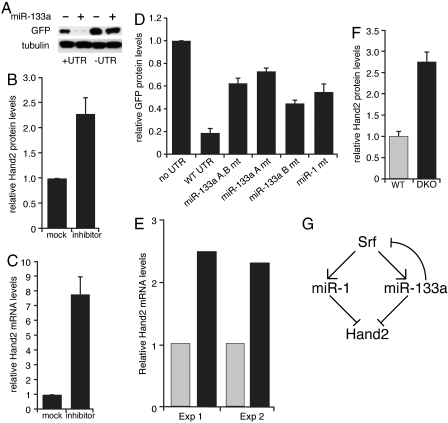
Fig. 5.
miR-133a regulation of Hand2 expression. (A) Western blot demonstrating that addition of miR-133a oligonucleotides to nondifferentiated C2C12 cells only inhibits GFP expression from reporter constructs containing the Hand2 3′UTR. Similar results were observed in three independent experiments. (B) miR-133a-blocking oligonucleotides increase endogenous Hand2 protein levels in primary cardiomyocytes. Hand2 protein levels were normalized to tubulin. (Mean ± SE are shown.) (C) miR-133a-blocking oligonucleotides increase Hand2 mRNA levels in primary cardiomyocytes, as determined by quantitative RT-PCR. Results shown represent three independent experiments. (Mean ± SE are shown.) (D) Mutation of miR-133a MREs relieves reporter repression in differentiated C2C12 cells. Effect of mutating the miR-1 MRE is shown for comparison. GFP protein levels quantified by densitometry were normalized to tubulin. Results represent six independent experiments. (Mean ± SE are shown.) (E) RT-PCR analysis of Hand2 mRNA levels in heart samples from miR-133a double knockout (DKO) mice and wild-type littermates. Five hearts at postnatal day 1 from each condition were used for each experiment. Gray bars represent wild type, black bars represent double knockouts. (F) Hand2 protein levels, relative to tubulin, in hearts from miR-133a DKO and wild-type littermates. Hearts from three wild-type and miR-133a DKO mice were analyzed. (Mean ± SE are shown.) (G) Model for miR-1 and miR-133a regulation of Srf and Hand2 expression, wherein partial loss of miR-133a feedback inhibition of Srf augments miR-1 response.
The affinity purification assays presented above suggested that the Hand2 3′UTR contains two distinct binding sites for miR-133a (Fig. 3 B and C). To test whether both mediate repression, we transfected the mutated GFP reporter constructs into C2C12 cells that had been differentiated to express miR-1 and miR-133a. Mutating either of the putative miR-133a MREs increased reporter gene expression, as compared to a reporter containing the wild-type 3′UTR, and mutating the miR-1 MRE increased GFP levels to about the same degree (Fig. 5D). Reporter expression was not as high as that observed when the Hand2 3′UTR was completely removed, however, suggesting that combinatorial actions of miR-1 and miR-133a, or perhaps additional microRNAs, might also contribute to repression.
The cardiac phenotype of mice deleted at both miR-133a loci (designated miR-133a double knockout) is similar in some respects to that of mice lacking miR-1 (17), so we next asked whether Hand2 expression levels were similarly elevated. Samples from five wild-type and five miR-133a-double knockout mice were pooled and analyzed by RT-PCR and Western blotting. Hand2 mRNA and protein levels were increased in the knockouts by about 2.5-fold (Fig. 5 E and F). A model describing the network of miR-1 and miR-133a interactions is presented in Fig. 5G.
Discussion
Identification of mRNA targets is a central problem in understanding how microRNAs contribute to gene regulation. Although there is a fairly high degree of discordance among the various prediction algorithms, there is agreement that individual transcripts can potentially be regulated by multiple microRNAs. The fact that target sites for distinct microRNAs are often conserved simultaneously also supports the concept of combinatorial regulation (21, 22). We developed an affinity purification assay to identify microRNAs associated with the Hand2 3′UTR and showed that miR-133a was a potential regulator of Hand2 expression. Further, our sequential affinity purification assay demonstrated that miR-1 and miR-133a can bind simultaneously to the Hand2 3′UTR and provided a mechanism for synergistic regulation. Using miR-133a specific microRNA inhibitors and mimics, we showed that miR-133a regulated Hand2 mRNA and protein expression. Additionally, we demonstrated that the loss of this microRNA was associated with an elevation in Hand2 expression in vivo.
Because miR-1 and miR-133a are generated from bicistronic precursors, their transcriptional regulation occurs in concert. miR-1 and miR-133a are believed to control the expression of distinct mRNA transcripts, however, and no common mRNA targets have been identified. Our findings indicate that Hand2 is repressed by both miR-1 and miR-133a. It is possible, therefore, that some of the shared features of the miR-1 and miR-133a knockouts are due to their coordinate regulation of Hand2 expression. These two microRNAs could interact with other shared targets as well and thereby coordinately control the expression of additional proteins involved in cardiac growth and/or cell survival. Nonetheless, the finding that miR-1 and miR-133a have opposing effects on apoptosis (23) indicates that at least some of their targets are distinct. Interestingly, the level of miR-133a binding to the Hand2 3′UTR, as measured in our affinity purification assay, is considerably greater than that observed for miR-1 (Fig. 2 A and B). This lesser degree of miR-1 interaction could explain why deleting one of the two miR-1 loci was sufficient to generate a phenotype, whereas this was not the case for miR-133a. An alternative explanation, diagrammed in Fig. 5G, is that the partial loss of miR-133a expression caused by the knockout of a single miR-133a gene might increase serum response factor (Srf) expression due to the loss of feedback inhibition. This could stimulate expression of the remaining miR-133a locus and increase levels of miR-1, both of which would reduce levels of Hand2. Thus, although loss of one miR-1 locus caused Hand2 levels to increase, deletion of a single miR-133a locus might activate a compensatory Srf response and not have this effect.
Generally, individual microRNA interactions have relatively modest effects, which has been interpreted as indicating that these molecules are primarily involved in “fine-tuning” (24, 25). These effects might be more prominent if other microRNAs were altered coordinately, and the affinity purification method should be useful for identifying candidates. Although this study focused on characterizing the role of miR-133a, we identified several other microRNAs associated with Hand2 using the multiplex assay and these may also be capable of regulating Hand2 expression. A multiplicity of interacting microRNAs could impart a wide variety of signaling inputs onto the regulation of Hand2 expression. Not all microRNAs are represented in the multiplex arrays, however, so it is possible that additional Hand2 regulators (false negatives) remain to be identified. Examination of the microRNAs that were identified is informative, however. For example, although miR-143 was predicted to regulate Hand2 (Fig. 1A) and is expressed in the heart (Fig. S1), it was not identified in our affinity purifications. A miR-143 inhibitor did not relieve repression of a luciferase reporter in cardiomyocytes (Fig. S3), and a mimic did not reduce expression in HEK293 cells, suggesting that this particular interaction does not represent a false negative. Further studies, perhaps examining other 3’UTR sequences, will be required to define the incidence of false negatives inherent to this approach. Additionally, it is important to consider that a negative interaction in one cell type could be positive in another. For example, we identified miR-146b as a Hand2 associated microRNA in cardiomyocytes, but this interaction was not detected in screens performed in HEK293 cells, despite the fact that miR-146b expression in the latter is considerably higher (Fig. S4). These observations further illustrate the importance of empirical assays for identifying microRNA interactions. The affinity purification method should add to the armamentarium of approaches for identifying such interactions. Unlike the HITS-CLIP (high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation) (26) or PAR-CLIP (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (27) methods, affinity purification identifies the interacting microRNA itself rather than an MRE that must subsequently be tested to determine its cognate microRNA. These methods should be complementary, however, and generally applicable for identifying other microRNA-mRNA interactions.
Methods
MS2 Pulldown Assay (Modified from Ref. 28).
HEK293 cells and C2C12 cells were transfected using the Lipofectamine reagent (Invitrogen). Primary rat cardiomyocytes were infected with lentivirus constructs. Transfected or infected cells were harvested 48–72 h postinfection by a wash with PBS followed by brief vortex and incubation on ice in lysis buffer [20 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.05% Igepal, 60 U/mL Superase-In (Ambion), 1 mM DTT, protease inhibitors (Roche)]. The lysates were precleared by centrifugation and then incubated with MBP-MS2 bound amylose beads for 3 h at 4 °C. The beads were subsequently washed with lysis buffer, and bound RNAs were purified using Trizol (Sigma). RT-PCR for GFP was performed to confirm purification of the GFP transcript prior to analysis of associated microRNAs. MicroRNAs purified with the MS2-tagged Hand2 3′UTR GFP transcript were identified using the ABI multiplex Taqman microRNA assay for rodent. Specificity of the identified microRNA interactions was confirmed by analysis of microRNAs associated with the MS2-tagged Hand2 3′UTR expressing virus or a negative control virus expressing the MS2-tagged reverse Hand2 3′UTR using Taqman microRNA assays.
Sequential Pulldown Assay.
Sequential pulldown assays were performed using primary rat cardiomyocytes infected with viruses expressing the MS2-tagged Hand2 3′UTR. Cells were transfected with biotinylated miR1 oligonucleotides. Following the MS2 pulldown, the RNA complexes bound to the amylose beads were eluted with lysis buffer containing 20 mM maltose. The eluates were then incubated with streptavidin beads (preblocked with yeast tRNA and BSA) (DynaL) for 3 h at 4 °C, washed, and RNA purified with Trizol. Mature microRNAs were detected using Taqman assays.
Supplementary Material
Acknowledgments.
We thank Ronald Kwong for technical assistance, members of the Goodman and Mandel labs for helpful comments, Greg Yochum and Xiaolu Cambronne for assistance with the biotinylated microRNA binding assays, and Gail Mandel and Soo Lee for critical reading of the manuscript and insightful discussions. This research was supported by grants from the National Institutes of Health.
Footnotes
Conflict of interest statement: Eric N. Olson holds equity in Miragen Therapeutics, which is developing miRNA-based therapies for muscle disease.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013162107/-/DCSupplemental.
References
- 1.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 2.Chen JF, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Lal A, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 13.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 20.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Chan CS, Elemento O, Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 24.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 25.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 30.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissuespecific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.