Abstract
Grain yield in many cereal crops is largely determined by grain size. Here we report the genetic and molecular characterization of GS3, a major quantitative trait locus for grain size. It functions as a negative regulator of grain size and organ size. The wild-type isoform is composed of four putative domains: a plant-specific organ size regulation (OSR) domain in the N terminus, a transmembrane domain, a tumor necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family cysteine-rich domain, and a von Willebrand factor type C (VWFC) in the C terminus. These domains function differentially in grain size regulation. The OSR domain is both necessary and sufficient for functioning as a negative regulator. The wild-type allele corresponds to medium grain. Loss of function of OSR results in long grain. The C-terminal TNFR/NGFR and VWFC domains show an inhibitory effect on the OSR function; loss-of-function mutations of these domains produced very short grain. This study linked the functional domains of the GS3 protein to natural variation of grain size in rice.
Keywords: grain weight, Oryza sativa L., protein domain, yield
In recent years, genes for grain and fruit sizes have been isolated from several plant species (1–8), providing opportunities for understanding genetic and molecular mechanisms regulating these traits. In rice, yield per plant is determined by three component traits: number of panicles (tillers) per plant, number of grains per panicle, and grain weight. Extensive genome mapping studies have identified hundreds of QTLs (quantitative trait loci) for yield traits (9). Although a number of QTLs for tillering (10), grain number (11, 12), grain size (8, 13–15), and panicle size and plant architecture (16–18) have been cloned, molecular characterization of these and many more genes is needed to understand the genetic and molecular bases of yield (9).
A major QTL for grain size (GS3) in rice was previously identified on chromosome 3 in a number of studies across different genetic backgrounds and environments (19–21). Fan et al. (8) identified the candidate gene for GS3. By comparative sequencing analysis, they found a nonsense mutation in the second exon of the putative GS3 shared among all of the large-grain varieties tested in comparison with varieties with smaller grains. This mutation caused a 178-aa truncation in the C terminus of the predicted protein, which was widespread in the global rice germplasm collections (22, 23), indicating that this mutation had an ancient origin and played an important role in grain size variation and domestication of the cultivated rice.
Here we report the genetic and molecular analysis of GS3, which revealed several important structural and functional features of the GS3 protein in grain size regulation.
Results
Validation of GS3 on Grain Size Regulation.
We validated the effect of GS3 on grain size by transformation. A construct CT9.8 (Fig. S1A), containing a 9.8-kb genomic DNA fragment encompassing GS3 amplified by PCR from rice cultivar Chuan 7 (short grain) in the binary vector pCAMBIA1301, was transformed into rice cultivar Minghui 63 (long grain). Thirty or more seeds were produced from 19 independent T0 transformants, and shorter and slightly wider grains were observed in all of the 19 plants (Fig. 1A). We then field-examined two T1 families randomly selected from T0 plants with single-copy transferred DNA insertion confirmed by Southern blot and observed perfect cosegregation between the transgene and the phenotype. All transgene-positive plants produced shorter and slightly wider grains than the negative segregants (Fig. S1 B–D). We further field-tested T3 homozygous lines derived from three independent single-copy transformants. Compared with the wild-type Minghui 63, all of the transgenic lines showed a highly significant decrease in grain length, together with a slight but significant increase in grain width, resulting in a large reduction in grain weight (Table 1).
Fig. 1.
Effect of GS3 on grain size and plant morphology exhibited by transgenics. (A–C) Grains of positive (Left) and negative (Right) segregants from T1 family by (A) transforming Minghui 63 with CT9.8 containing the genomic fragment of Chuan 7; (B) transforming Chuan 7 with the RNAi-GS3 construct; and (C) overexpressing GS3 using the 35SFL construct. (D–F) Positive (Left) and negative (Right) segregants from T1 family overexpressing GS3 using the 35SFL construct: (D) whole plants; (E) panicles and flag leaves; and (F) main culms, with arrowheads indicating the internodes.
Table 1.
Comparison of grain traits of three independent homozygous transgenic lines with Minghui 63
| Trait | No. of plants | Minghui 63 | C6 | C10 | C15 |
| Grain length (mm) | 10 | 9.93 ± 0.03 | 7.69 ± 0.03 | 7.61 ± 0.05 | 7.83 ± 0.05 |
| P | 1.09 × 10−21 | 1.92 × 10−19 | 2.31 × 10−19 | ||
| Grain width (mm) | 10 | 2.87 ± 0.01 | 3.09 ± 0.01 | 3.09 ± 0.02 | 3.07 ± 0.02 |
| P | 1.71 × 10−11 | 3.72 × 10−10 | 2.51 × 10−9 | ||
| 1,000-grain weight (g) | 30 | 25.73 ± 0.15 | 18.38 ± 0.15 | 18.24 ± 0.25 | 19.85 ± 0.10 |
| P | 2.47 × 10−41 | 7.50 × 10−34 | 4.73 × 10−40 |
All data are given as mean ± SEM. Each P value for each trait was obtained from a t test between Minghui 63 and the transgenic line.
We also used RNA-interference (RNAi) to suppress the expression of GS3 in Chuan 7 (Fig. S1A). Because it is very difficult to transform Chuan 7 (an indica cultivar), we obtained only two positive transgenic plants, both of which showed significantly increased grain length (Fig. 1B), without obvious change in plant morphology. One of the two T1 families was analyzed for cosegregation between the grain traits and the transgene (Fig. S1E and Table S1). The grain length of the positive segregants was longer than that of negative plants. The correlation between grain length and GS3 expression level was −0.78, significant at P < 0.01 (n = 14). In addition, the grain weight of the positive plants also significantly increased compared with the negative segregants.
Overexpression of GS3 Conferred Pleiotropic Effects to Rice Plants.
To investigate possible effect of GS3 at the whole-plant level, we overexpressed the gene by transforming Minghui 63 with a construct 35SFL (Fig. S1A) containing the full-length cDNA (fl-cDNA, Osigcea013f09t3) of GS3 from the plumule of an indica rice cultivar, Guangluai 4 (medium grain) (8). Twenty-three independent transformants were obtained, each with 30 or more seeds. Overexpressing GS3 greatly reduced plant size, including shortened height, leaves, and panicles, in addition to short grains (Fig. 1 C–F). We field-tested two T1 families randomly selected from T0 plants. All of the positive segregants showed reduced grain length compared with negative segregants (Fig. S1 F–H). Two homozygous transgenic lines were further compared with the wild-type Minghui 63 for yield traits by field test. The transgenic lines showed a large reduction in grain length and weight, as well as number of grains per panicle, with a significant increase in tillers, whereas GS3 overexpression did not affect flowering time (Table S2).
Plant-Specific Unique Domain in the GS3 Protein.
The GS3 protein was previously predicted as consisting of four putative domains (8): a phosphatidylethanolamine-binding protein (PEBP)-like domain of 54 aa at the N terminus, a transmembrane region at amino acids 97–117, a TNFR (tumor necrosis factor receptor)/NGFR (nerve growth factor receptor) family cysteine-rich domain at sites 116–155, and a von Willebrand factor type C (VWFC) 60–80 aa in length in the C-terminal region. Later on, however, the PEBP domain was no longer predicted by the same analysis of the GS3 sequence (http://www.ebi.ac.uk/Tools/InterProScan/), whereas the other domains still remained. Moreover, GS3 was not identified in the recent analyses of rice PEBP protein family by other groups (24, 25). We performed an alignment analysis of the previously identified putative PEBP domain of GS3 with the 19 proteins identified as PEBP family members (24) and found that the putative GS3 PEBP was only approximately one third of the length of the predicted PEBP domain in those proteins, sharing poor similarity (20.3–28.4%) in the PEBP region. Thus the putative PEBP domain is not found in GS3.
We searched databases for homologous sequences of the GS3 N-terminal region and found an N-terminal motif 66 aa in length (sites 7–72) of GS3 not reported as a conserved domain in the literature but sharing high similarity with the N-terminal regions of proteins from a number of plants, ranging from angiosperm to gymnosperm (Fig. S2), including DEP1 identified as a gene for panicle architecture in rice (17). Interestingly, a GS3 ortholog in maize was found to be involved in kernel development (26). GS3 exhibited the highest similarity with Zea mays ZmGS3-1 (95.8%) and the lowest with Arabidopsis AtGS3 (49%) in this region. However, this domain was not found in animals or other organisms, thus is highly likely plant specific. To reflect the function of this protein domain (see below), we tentatively named it the organ size regulation (OSR) domain.
Unique Allele from Chuan 7 Had a Strong Function in Grain Size Regulation.
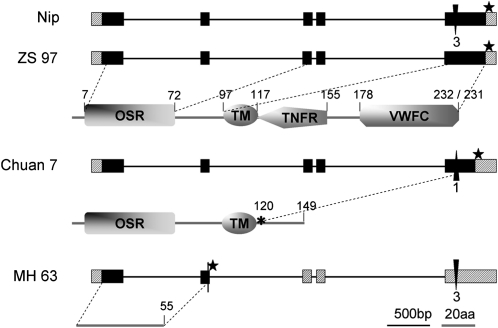
The grains of Chuan 7 are lighter than other varieties, primarily because of shorter length. For example, the 1,000-grain weight of Minghui 63 is usually 26–28 g, and that of Zhenshan 97 is 22–24 g, whereas 1,000 grains of Chuan 7 only weigh 12.5 g (8, 19, 20, 27). Grains of transgenic plants harboring the Chuan 7 genomic segment (CT9.8) in Minghui 63 background obtained in this study were also smaller than those of Zhenshan 97, although larger than Chuan 7 (Table 1 and Table S3). Such grain size differences indicate that GS3 encoded by Chuan 7 may be a stronger allele than that of Zhenshan 97 in negatively regulating grain size. We thus conducted comparative sequencing of the entire GS3 region of several varieties (Fig. 2): Minghui 63 (long grain), Zhenshan 97 and Nipponbare (medium grain), and Chuan 7 (short grain). The predicted protein of Zhenshan 97, whose coding region was identical to Guangluai 4, was 231 aa in length. Relative to Zhenshan 97, Minghui 63 had a C→A substitution at 165 bp downstream from the predicted translation start site (ATG), causing premature termination of the predicted protein and resulting in long grain, which was also observed in several studies (8, 22, 23). Chuan 7 had a 1-bp deletion at 357 bp (C) downstream from the predicted translation start site, which caused a frameshift mutation in the C terminus that yielded a truncated 149-aa protein (Fig. 2). Interestingly, Nipponbare had a 3-bp (TCC) insertion at the same site, resulting in an in-frame addition of a serine residue. Therefore, GS3 has at least four different alleles according to the coding sequence: GS3-1 (Zhenshan 97), GS3-2 (Nipponbare), GS3-3 (Minghui 63), and GS3-4 (Chuan 7).
Fig. 2.
Comparison of the genomic DNA and predicted protein sequences of GS3 in Zhenshan 97 (ZS97, GS3-1), Nipponbare (Nip, GS3-2), Minghui 63 (MH63, GS3-3), and Chuan 7 (GS3-4), relative to ZS97. Genomic DNA: hatched boxes represent 5′ and 3′ UTR, and five exons are indicated by black boxes. The thin lines between black boxes represent introns. Black triangles represent a 3-bp insertion (Nip) and a 1-bp deletion (Chuan 7). Stop codons are marked by stars. Protein structure (underneath each genomic DNA fragment): the domains encoded by Nip/ZS97 allele are shown, with the numbers indicating the positions of the domains. The 1-bp deletion in Chuan 7 causes frameshift from site 120 (marked with asterisk), resulting in the loss of TNFR and VWFC domains. The putative protein encoded by Minghui 63 allele contains no functional domain because of premature termination.
To investigate the effects of these variants and their abundance in the rice germplasm, we sequenced the GS3 genomic region and measured grain length for a total of 82 accessions, mostly from a core collection (28) of the Chinese cultivated rice germplasm (Table S3). Eleven of the 82 accessions contained the C→A substitution as in Minghui 63; all of them produced long grain. The remaining 70 accessions (with Chuan 7 excluded) were resolved into two distinct groups using STRUCTURE analysis (29) based on data of 16 simple sequence repeat markers, essentially an indica group and a japonica group (Table S3). There was no significant difference in grain length between the two genotypes (TCC insertion/deletion) within the groups (Table S4), suggesting that it is unlikely that this insertion/deletion has a large effect on grain size, although the numbers of accessions were small in two of the classes.
The GS3-4 allele encoded by Chuan 7 occurred only once in the 82 accessions (Table S3). It was neither found with additional sequences of 60 accessions of Oryza rufipogon and Oryza nivara from Bangladesh, China, India and Myanmar, Nepal, Sri Lanka, and Thailand, nor observed among the 54 diverse accessions published previously (22). Database searching of the predicted polypeptide sequence of the frameshift C terminus found no match with any conserved motif.
We evaluated the effects of these alleles with near isogenic lines (NILs) in the background of Minghui 63: NIL(zs97) carrying an introgressed GS3-1 from Zhenshan 97, and NIL(c7) with the GS3-4 from Chuan 7 (Fig. S3A). NIL(zs97) was obtained by three successive backcrosses, using Minghui 63 as the recurrent parent, with the F1 from a cross between Minghui 63 and RIL236, a recombinant inbred line containing 55% of Minghui 63 background from a cross between Zhenshan 97 and Minghui 63. NIL(c7) was obtained by six backcrosses, using Minghui 63 as the recurrent parent, with the F1 from a cross between Minghui 63 and Chuan 7. The two NILs were crossed, and self-pollination of the F1 produced three genotypes: NIL(c7), NIL(zs97), and NIL(het). As expected, NIL(c7) produced the shortest grains, and grains of NIL(zs97) were also significantly shorter than Minghui 63 (Table S5). Grain length of the heterozygote, NIL(het), was not significantly different from the mean of the two homozygotes (Fig. S3B and Table S5), indicating that there is no genetically dominant–recessive relation between the two alleles. Grain weight essentially followed the same trend, which was also reflected in grain yield per plant. Small but statistically detectable differences were observed among the four genotypes in number of grains per panicle and number of tillers per plant (Table S5).
Expression Pattern of GS3.
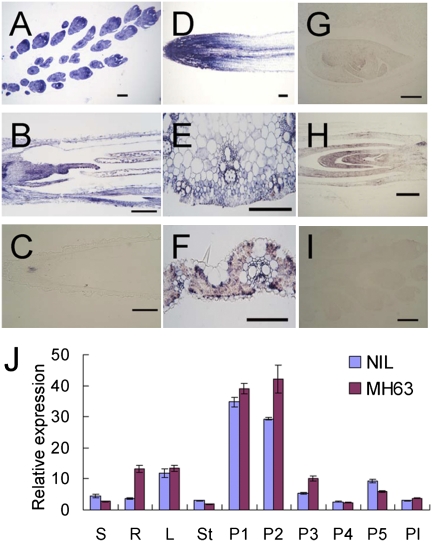
RNA in situ hybridization showed that GS3 was highly expressed in young panicle, and the signal gradually decreased with panicle development (Fig. 3 A–C). Weak signals were observed in other tested tissues, including embryo, shoot apical meristem, leaf, and stem (Fig. 3 E–H). Interestingly, GS3 was also highly expressed in root tips (Fig. 3D). Real-time PCR analysis of Minghui 63 and NIL(c7) showed that GS3 mRNA preferentially accumulated in panicles less than 5 cm in length in both genotypes, whereas the transcript level was low in other tissues assayed (Fig. 3J). This observation was consistent with the result from in situ hybridization analysis. These suggested that the expression level and pattern were not the cause of the loss of function of GS3 in Minghui 63.
Fig. 3.
Expression pattern of GS3. (A–H) Expression of GS3 in various tissues revealed by in situ hybridization: (A) panicle at stage of pollen mother cell formation, (B) panicle at pollen mother cell meiosis, (C) panicle at pollen grain filling, (D) root at trefoil stage, (E) stem at heading stage, (F) leaf at tillering stage, (G) endosperm and embryo 14 d after flowering, and (H) longitudinal section of 7-d-old seedling. (I) Negative control by hybridizing the young panicle with the sense probe. All tissues were sampled from Zhenshan 97 (medium grain). (Scale bars, 200 μm.) (J) Comparative expression pattern of GS3 in Minghui 63 and NIL(c7) in various organs using real-time RT-PCR analysis: S, seedling at trefoil stage; R, root at trefoil stage; L, leaf at tillering stage; St, stem at heading stage; P1, young panicle <1 cm in length; P2, young panicle <5 cm in length; P3, young panicle 5–10 cm in length; P4, panicle before heading >10 cm in length; P5, panicle 5 d after heading; Pl, plumule.
Differential Functions of the Domains.
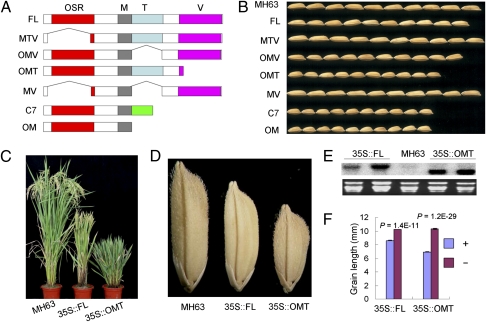
The fact that Minghui 63 with premature termination in the OSR domain produced long grain, whereas the frameshift mutant lacking TNFR and VWFC domains produced the shortest grain, suggested that these domains may exert antagonistic effects in grain size regulation. To investigate the roles of these domains in regulating grain size, seven constructs were made by domain deletions (Fig. 4A), in which the coding sequences were driven by the GS3 native promoter from Chuan 7 and again delivered to Minghui 63. On the basis of grain length of field-planted T1 plants (Fig. 4B and Table 2), it is clear that the OSR domain was both necessary and sufficient for reducing grain size; all of the constructs with this domain produced significantly shorter grains, whereas constructs without this domain did not significantly affect grain size. The construct of fl-cDNA (GS3-1) had the mildest effect on grain size reduction (9.5%), whereas the one (OM) with only the OSR domain had the strongest effect (29.1%), even larger than that of C7 (24.5%). Thus the TNFR and VWFC domains could to some extent inhibit the effect of OSR in grain size reduction. Moreover, the frameshift C terminus encoded by GS3-4 allele also seemed to have some effect on grain size regulation. Comparison of numerical values of size reduction indicated that the VWFC domain seemed to have a larger effect of inhibiting OSR than did TNFR and that these two domains added to each other in regulating OSR for grain size.
Fig. 4.
Effects on grain size of the various domains assessed using transgenics. (A) Schematic representation of the coding sequences of the constructs with one or more domains deleted; (B) grains of the T1 generation transgenic plants compared with Minghui 63. OSR, organ size regulation domain; M, putative transmembrane domain; T, TNFR/NGRF domain; V, VWFC domain. Green box represents the polypeptide sequence resulting from frameshift. (C and D) Effect of VWFC in inhibiting OSR in regulating growth of plant and grain size. From left to right: Minghui 63, transgenic plants overexpressing the fl-cDNA (FL), or the truncated cDNA (OMT). (E) Northern blot analysis of GS3 transcripts in wild-type plants and in two positive segregants from respective T1 generations. (F) Grain length of T1 plants overexpressing fl-cDNA (FL) or the truncated cDNA (OMT). + and −, positive and negative segregants. Data are given as mean ± SEM. Student's t test was used to generate the P values.
Table 2.
Grain length of transgene-positive and transgene-negative segregants in one of the T1 families for each construct
| Constructs | Positive (mm) | Negative (mm) | P | Reduction (%)* |
| C7 | 7.35 ± 0.07 (n = 16) | 9.74 ± 0.03 (n = 14) | 1.48 × 10−22 | 24.5 |
| FL | 9.05 ± 0.02 (n = 23) | 10.00 ± 0.04 (n = 7) | 1.12 × 10−18 | 9.5 |
| OMT | 7.96 ± 0.03 (n = 21) | 9.95 ± 0.06 (n = 8) | 8.83 × 10−23 | 20.0 |
| OMV | 8.77 ± 0.03 (n = 20) | 10.23 ± 0.04 (n = 9) | 1.72 × 10−21 | 14.3 |
| OM | 7.12 ± 0.07 (n = 25) | 10.05 ± 0.10 (n = 5) | 5.50 × 10−17 | 29.1 |
| MTV | 9.66 ± 0.04 (n = 21) | 9.77 ± 0.03 (n = 9) | 0.0520 | |
| MV | 9.79 ± 0.05 (n = 23) | 9.87 ± 0.08 (n = 11) | 0.1656 |
All data are given as mean ± SEM. P values were obtained by t tests between the positive and negative plants in the same family.
*Extent of grain length reduction of the transgene positive plants relative to the negative plants.
To further evaluate the effect of the VWFC domain, we overexpressed a truncated cDNA sequence (OMT) with the VWFC domain deleted. Transformants produced much shorter grains and highly dwarfed plants compared with those overexpressing the fl-cDNA (Fig. 4 C–F), providing further evidence that VWFC has a general role of inhibiting the effect of OSR in regulating plant growth and grain size.
Discussion
This study has revealed several structural and functional features of the GS3 protein in grain size regulation. First, the plant-specific domain OSR identified in the N terminus is the most important one, which is both necessary and sufficient for GS3 to function as a negative regulator of grain size as well as organ size. Loss of function or deletion of this domain would result in long grain. Second, there is an intramolecular interaction between the OSR domain and the C-terminal domains, TNFR/NGFR and VWFC. The C-terminal domains inhibit the function of OSR on grain and organ size reduction, independently of tissues or cell types. This phenomenon is similar to previously reported autoregulation in the plant steroid receptor BRI1 (30) and autoinhibition in the human platelet-derived growth factor β-receptor (31), both of which were membrane proteins and whose functions involved kinase activity. However, the biochemical function of the GS3 protein remains to be determined.
Because grain size has been the target of both natural selection and breeding (32), the GS3 locus provides an informative system for studying the evolutionary processes underlying rice domestication and breeding. The GS3-1 allele is probably the prototype and corresponds to medium grain length, which is the genotype for the majority of widely grown medium grain indica varieties (e.g., Zhenshan 97). Relative to GS3-1, an in-frame insertion of 3 bp (GS3-2) in the fifth exon did not change the grain length, which is the case for most of the temperate japonica varieties (e.g., Nipponbare). One base pair deletion at the same site caused frameshift of the C-terminal domains of the protein and thus generated an allele (GS3-4) with very strong function as a negative regulator producing small grain. However, this allele is extremely rare in rice germplasm collections, indicating that such small grain has not been favored by natural selection or artificial breeding. In contrast, premature termination of the OSR domain by a single base substitution in the second exon greatly increased grain length, which is the case for long grain varieties widely cultivated in the world, suggesting that this allele has been highly favored in breeding programs. These results are directly useful in breeding programs to modify grain size and increase grain yield using both molecular marker-assisted selection and genetic transformation techniques (33, 34).
Methods
Field Growth of Rice Plants.
The rice plants examined under natural field conditions were grown in normal rice growing seasons at the Experimental Station of Huazhong Agricultural University, Wuhan, China. Seeds were planted in a seed bed in mid-May and transplanted to the field in mid-June. The planting density was 16.5 cm between plants in a row, and the rows were 26 cm apart. Field management, including irrigation, fertilizer application, and pest control, followed essentially the normal agricultural practice.
Measurements of Grain Traits.
Harvested paddy rice was air-dried and stored at room temperature before testing. Ten randomly chosen, fully filled grains from each plant were lined up length-wise along a vernier caliper to measure grain length and then arranged by breadth to measure grain width. Grain weight was calculated on the basis of 200 grains and converted to 1,000-grain weight.
Generation of Constructs and Transformation.
For preparing the GS3 construct of the genomic DNA from Chuan 7, a 9,868-bp fragment spanning from 2,780 bp upstream of the translation start codon to 1,371 bp downstream of the termination codon of GS3 was amplified by two-step PCR from rice cultivar Chuan 7 with two pairs of primer, 1512f–9247r and U9EcoRIF–RBP (Table S6). The overlapped region of the two PCR products contained an EcoRI site, which was used to cut and ligate the two fragments. The fragment was cloned into the binary vector pCAMBIA1301.
Two cDNA clones were used for the overexpression constructs. The first was the fl-cDNA (Osigcea013f09t3) from the plumule of Guangluai 4, cloned in pBluescript SK2 (Stratagene). The second was the fl-cDNA with 3′-deletion prepared by PCR from the full-length cDNA with primers F1SacI and VWFCKpnI (Table S6). These two clones were digested with SacI and KpnI and inserted into pCAMBIA1301s under the control of the Cauliflower mosaic virus CaMV-35S promoter.
To construct an RNAi vector for GS3, a 790-bp fragment of GS3 was digested with BamHI and KpnI from the above-mentioned fl-cDNA clone and inserted into the pDS1301 vector, which was a modified version of pCAMBIA1301 (35).
All of these constructs were independently introduced into the Agrobacterium strain EHA105, and transformation was done as described previously (36).
DNA Blot Analysis.
The experimental procedures for Southern blot analysis, including DNA extraction, enzyme digestion, electrophoresis, and hybridization, were essentially as described previously (37). The PCR product of GUS (β-glucuronidase) gene using GUS1.6F and GUS1.6R as primers was purified and prepared as probe for Southern blot analysis.
Expression Analyses.
We isolated total RNA from the tissues/organs using an RNA extraction kit (TRIzol reagent; Invitrogen) according to the manufacturer's instructions. For quantitative real-time PCR, approximately 3 μg total RNA was reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen) in a volume of 100 μL to obtain cDNA. A pair of primers, G53F and G53R, was used to amplify the transcript of GS3, with Actin1F and Actin1R for β-Actin1 as the internal control (Table S6). We carried out quantitative real-time PCR in a total volume of 25 μL containing 2 μL of the reverse-transcribed product above, 0.25 mM gene-specific primers, and 12.5 μL SYBR Green Master Mix (Applied Biosystems) on an Applied Biosystems 7500 Real-Time PCR System according to the manufacturer's instructions. The measurements were obtained using the relative quantification method (38).
For Northern blot analysis, 15 μg of total RNA from leaves was separated on 1.2% (wt/vol) denaturing agarose gel and transferred to nylon membrane. A GS3 fragment amplified with gene-specific primers CFPXbaIF and F-ExHindIIIR (Table S6) was labeled with 32P-dCTP using Random Primer kit (Invitrogen) and hybridized to the RNA blots.
In Situ Hybridization.
The GS3 probe was amplified with the full-length cDNA clone as the template using the gene-specific primers F1SacI and VWFCKpnI (Table S6). A 485-bp PCR fragment was then inserted into the pGEM-T vector (Promega) for RNA transcription. The sense and antisense RNA probes were produced by T7 and SP6 transcriptase labeled with digoxigenin (Roche), respectively. RNA in situ hybridization and immunological detection were done as described previously (39).
Supplementary Material
Acknowledgments
We thank Dr. Joe M. Jez for comments on the manuscript. This work was supported by the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the 948 Project of the Ministry of Agriculture, the 111 Project, and the National Natural Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014419107/-/DCSupplemental.
References
- 1.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 2.Frary A, et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 3.Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008;22:1331–1336. doi: 10.1101/gad.463608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, et al. Cell Number Regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell. 2010;22:1057–1073. doi: 10.1105/tpc.109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan C, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 9.Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 10.Li X, et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- 11.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 12.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 13.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 14.Song X, Huang W, Shi M, Zhu M, Lin H. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 15.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 16.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 18.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 19.Yu SB, et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing Z, et al. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet. 2002;105:248–257. doi: 10.1007/s00122-002-0952-y. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Thomson M, McCouch SR. Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics. 2004;168:2187–2195. doi: 10.1534/genetics.104.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takano-Kai N, et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009;182:1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan C, Yu S, Wang C, Xing Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet. 2009;118:465–472. doi: 10.1007/s00122-008-0913-1. [DOI] [PubMed] [Google Scholar]
- 24.Chardon F, Damerval C. Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol. 2005;61:579–590. doi: 10.1007/s00239-004-0179-4. [DOI] [PubMed] [Google Scholar]
- 25.Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, et al. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor Appl Genet. 2010;120:753–763. doi: 10.1007/s00122-009-1196-x. [DOI] [PubMed] [Google Scholar]
- 27.Hua J, et al. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics. 2002;162:1885–1895. doi: 10.1093/genetics/162.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, et al. A core collection and mini core collection of Oryza sativa L. in China. Theor Appl Genet. 2010 doi: 10.1007/s00122-010-1421-7. 10.1007/s00122-010-1421-7. [DOI] [PubMed] [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Chiara F, Bishayee S, Heldin CH, Demoulin JB. Autoinhibition of the platelet-derived growth factor beta-receptor tyrosine kinase by its C-terminal tail. J Biol Chem. 2004;279:19732–19738. doi: 10.1074/jbc.M314070200. [DOI] [PubMed] [Google Scholar]
- 32.Gegas VC, et al. A genetic framework for grain size and shape variation in wheat. Plant Cell. 2010;22:1046–1056. doi: 10.1105/tpc.110.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittler R, Blumwald E. Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol. 2010;61:443–462. doi: 10.1146/annurev-arplant-042809-112116. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q. Strategies for developing Green Super Rice. Proc Natl Acad Sci USA. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan B, Shen X, Li X, Xu C, Wang S. Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta. 2007;226:953–960. doi: 10.1007/s00425-007-0541-z. [DOI] [PubMed] [Google Scholar]
- 36.Ge X, Chu Z, Lin Y, Wang S. A tissue culture system for different germplasms of indica rice. Plant Cell Rep. 2006;25:392–402. doi: 10.1007/s00299-005-0100-7. [DOI] [PubMed] [Google Scholar]
- 37.Liu K, et al. A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor Appl Genet. 1997;95:809–814. [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Dai M, Hu Y, Zhao Y, Liu H, Zhou D. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007;144:380–390. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.