Abstract
During recent years, increased efforts have focused on elucidating the secretory function of skeletal muscle. Through secreted molecules, skeletal muscle affects local muscle biology in an auto/paracrine manner as well as having systemic effects on other tissues. Here we used a quantitative proteomics platform to investigate the factors secreted during the differentiation of murine C2C12 skeletal muscle cells. Using triple encoding stable isotope labeling by amino acids in cell culture, we compared the secretomes at three different time points of muscle differentiation and followed the dynamics of protein secretion. We identified and quantitatively analyzed 635 secreted proteins, including 35 growth factors, 40 cytokines, and 36 metallopeptidases. The extensive presence of these proteins that can act as potent signaling mediators to other cells and tissues strongly highlights the important role of the skeletal muscle as a prominent secretory organ. In addition to previously reported molecules, we identified many secreted proteins that have not previously been shown to be released from skeletal muscle cells nor shown to be differentially released during the process of myogenesis. We found 188 of these secreted proteins to be significantly regulated during the process of myogenesis. Comparative analyses of selected secreted proteins revealed little correlation between their mRNA and protein levels, indicating pronounced regulation by posttranscriptional mechanisms. Furthermore, analyses of the intracellular levels of members of the semaphorin family and their corresponding secretion dynamics demonstrated that the release of secreted proteins is tightly regulated by the secretory pathway, the stability of the protein, and/or the processing of secreted proteins. Finally, we provide 299 unique hydroxyproline sites mapping to 48 distinct secreted proteins and have discovered a novel hydroxyproline motif.
The skeletal muscle is a highly dynamic organ responsible for locomotion and generation of body heat and plays an essential role in the maintenance of metabolic homeostasis. Furthermore, it is the major target for insulin-induced glucose uptake and has a high capacity to metabolize fatty acids. Impaired glucose metabolism and lipid metabolism in the skeletal muscle are characteristic hallmarks of insulin resistance associated with different diseases such as obesity, type 2 diabetes, and metabolic syndrome (1). During the last decade, data have emerged demonstrating that the skeletal muscle also plays an active role as an endocrine organ. Skeletal muscles have been suggested to be a source of secreted proteins, conceptualized as myokines that can influence metabolism and other biological processes in a systemic manner at different tissue targets. Regular exercise has many beneficial effects on whole body well-being: it improves insulin sensitivity, metabolic processes, and blood pressure and reduces inflammation mediated through alterations of gene and protein expression in different cell types. Several studies have described the association of physical inactivity with the development of different pathologies, including mental diseases as well as obesity, type 2 diabetes, metabolic syndrome, and cardiovascular diseases (1). It has been suggested that the skeletal muscle, through secreted molecules, could be transmitting many of the beneficial effects of physical activity by affecting whole body homeostasis in an autocrine, paracrine, and/or endocrine fashion.
The initial recognition of skeletal muscle as an endocrine organ originates from studies that identified interleukin (IL)-6 expression and secretion by contracting skeletal muscle. The plasma level of muscle-derived IL-6 increases in response to physical activity, and the secreted IL-6 can affect metabolic and inflammatory processes (2, 3). In addition to IL-6, other potential myokines, with either systemic effects or local effects in an autocrine and paracrine manner, have been identified, including IL-8 and IL-15 (3). Furthermore, a mouse model of inducible muscle hypertrophy was used as a tool for the identification of new myokines, resulting in the identification of fibroblast growth factor (FGF)-21 and follistatin-like 1 as secreted skeletal muscle proteins involved in metabolic processes and vascularization (4, 5).
Despite recent discoveries, the secretome of skeletal muscle has not been fully characterized, and the number of identified secreted proteins is still limited. The majority of studies investigating the secretory profile of the skeletal muscle have used traditional biochemical and molecular biology strategies to identify muscle-secreted proteins; although powerful, these approaches are restricted to a few proteins. Existing mass spectrometry-driven attempts to target the muscle-specific secreted proteome resulted in a limited number of identified secreted proteins (6–8). Comprehensive quantitative analyses of the muscle secretome could provide greater insight into muscle biology and the muscle-dependent cross-talk with other tissues. The fast development of quantitative mass spectrometry-based proteomics has allowed the qualitative as well as the quantitative evaluation of complex biological processes in a large scale manner (9). The SILAC1 methodology represents a powerful quantitative tool to study the dynamics of different biological processes, including in-depth characterization of the signaling cascades involved in various types of cellular differentiation (10–12).
Here we used triple encoding SILAC to identify and quantitatively evaluate the dynamics of secreted proteins during the differentiation process of the murine C2C12 skeletal muscle cell line. We identified 635 proteins that are secreted from skeletal myoblasts; to our knowledge, this is the largest data set covering the muscle secretome. Furthermore, we quantitatively profiled the secretion of 624 of these proteins during the course of skeletal muscle differentiation. Proteins previously known to be secreted from skeletal muscle were identified as well as many proteins not previously shown to be secreted by skeletal myoblasts. Based on gene ontology (GO) annotations, the identified secreted proteins are involved in various biological processes and molecular function categories, highlighting the diversity of the muscle secretome. Moreover, 188 secreted proteins were significantly and dynamically regulated during skeletal myogenesis, suggesting involvement in skeletal muscle development in either an autocrine or paracrine manner. Among the regulated secreted factors, we identified a family of proteins, the semaphorins, that are involved in muscle development. We investigated more closely the expression profiles of semaphorins at the mRNA and protein levels during the myoblast conversion. Finally, our approach resulted in the identification of 299 posttranslationally modified sites on 48 secreted proteins containing hydroxyproline residues and revealed a new motif distinct from the canonical hydroxyproline motif. To our knowledge, this also represents the largest data set of proline hydroxylation on secreted proteins.
EXPERIMENTAL PROCEDURES
Cell Culture
Murine C2C12 cells (American Type Culture Collection (ATCC)) were cultured in custom prepared DMEM containing 4.5 g/liter glucose, deficient in arginine and lysine (Invitrogen), and supplemented with 10% dialyzed fetal bovine serum (dFBS) (Invitrogen), 1000 IU/ml penicillin, 1000 IU/ml streptomycin, and 2 mm l-glutamine at 37 °C in humidified air containing 10% CO2. Regular l-Arg (Arg0) and l-Lys (Lys0) were added to the cell population to be harvested on day 0, whereas the cell populations harvested on days 2 and 5 of differentiation received different isotopic forms of l-arginine and l-lysine (Cambridge Isotope Laboratories and Sigma-Aldrich): day 2 cells, l-[13C6,14N4]arginine (Arg6) and l-[2H4]lysine (Lys4); and day 5 cells, l-[13C6,15N4]arginine (Arg10) and l-[13C6,15N2]lysine (Lys8). Cells were passaged at least five times to ensure complete incorporation of the labeled amino acids before differentiation (13). Differentiation was induced upon confluence by reducing the amount of dFBS to 2% with medium changes every 2 days.
Collection and Preparation of Conditioned Media (CM) for Analysis by Mass Spectrometry
Cells were passaged and differentiated as described above. On days 0, 2, and 5 of differentiation, cells were washed six times in serum-free media to reduce the amount of contaminating serum proteins and incubated for 12 h in corresponding serum-free labeling media. At the end of starvation, the CM were collected, centrifuged, and filtered using a 0.2-μm filter to ensure removal of any dead cells. The protein content of the individual media was measured using the Coomassie Plus reagent from Pierce. CM obtained from the three different time points were mixed in a 1:1:1 ratio according to the measured protein concentrations, and the combined media were concentrated using spin columns with a cutoff of 3000 Da (Vivaspin, Sartorius). The concentrated media were analyzed by a Novex 4–12% Bis-Tris gradient gel (Invitrogen), and proteins were visualized using Colloidal Blue (Invitrogen). Whole gel lanes of CM were cut into slices and subjected to in-gel digestion (14) with minor modifications. In-gel reduction was done for 45 min at 56 °C using 10 mm DTT in 50 mm ammonium bicarbonate followed by in-gel alkylation using 55 mm iodoacetamide in 50 mm ammonium bicarbonate for 30 min in the dark. Digestion was performed overnight with 12.5 ng/μl sequencing grade modified trypsin (Promega) at 37 °C. Extracted peptide mixtures were desalted and concentrated via StageTips essentially as described (15).
Mass Spectrometry and Data Analysis
Acidified peptide mixtures were separated by on-line C18 reverse-phase nanoscale liquid chromatography and analyzed by tandem mass spectrometry (LC-MS/MS). Reverse phase nanoscale LC was carried out by an Agilent 1200 nanoflow system (Agilent Technologies) connected to an LTQ-Orbitrap XL mass spectrometer (ThermoFisher) equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark). Full-scan MS spectra (m/z 300–2000) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400. The six most abundant ions in the linear ion trap were sequentially selected for sequencing by collision-induced dissociation using a collision energy of 35%. Ions already selected for fragmentation were dynamically excluded for 45 s. The lock mass calibration feature was enabled to improve mass accuracy (16). Additional mass spectrometric parameters included a spray voltage of 2.3 kV, no sheath and auxiliary gas flow, and a temperature of the heated capillary of 170 °C.
Acquired raw mass spectral data were processed and quantitated using MaxQuant v1.0.12.5 essentially as described (17). Briefly, a peak list was generated using the Quant element of MaxQuant using the following parameters: Orbitrap/FT Ultra; SILAC triplets with medium labels Arg6 + Lys4 and heavy labels Arg10 + Lys8; maximum of three labeled amino acids; maximum mass deviation allowed for precursor ions of 5 ppm and 0.5 Da for MS/MS events; and six most intense MS/MS peaks per 100 Da. A maximum of three missed cleavages was allowed, and enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline and between aspartic acid and proline. In addition, carbamidomethyl (Cys) was chosen as a fixed modification, and variable modifications included oxidation (Met), acetylation (protein N terminus), deamidation (NQ), Gln → pyro-Glu (N-terminal Gln), and oxidation (Pro). The MaxQuant-generated peak list was searched via the Mascot search engine v2.2 (Matrix Science, London, UK) against the mouse International Protein Index (IPI) database v3.44 containing 110,461 protein sequences, including a list of common contaminants such as keratins, serum albumin, trypsin, and concatenated with reverse copies of all sequences. Subsequently, the acquired Mascot dat files in association with raw files were processed and quantitated applying the Identify element of MaxQuant using the following parameters: peptide and protein false discovery rate, 0.01; maximum peptide posterior error probability, 1; minimum peptide length, 6; and minimum unique peptides and minimum peptides, 1. Razor and unique peptides, including unmodified and all modified peptides except oxidation (Pro) identified peptides, were subjected to quantitation with a minimum ratio count of 1. Only proteins identified with at least two peptides and one unique peptide were included in the final list of identified proteins. We used ProteinCenter v2.0 (Proxeon Biosystems), a proteomics data mining and management software, to filter our large data set of proteins to isolate secreted proteins. Proteins were classified as secreted based on the predicted presence of a signal peptide in the identified protein combined with extracellular filtering based on cellular component information from GO annotations (18).
Bioinformatics Analyses
To extract secreted proteins, protein identifications were filtered using ProteinCenter (Proxeon Biosystems). The entire IPI mouse database (version 3.52) was subjected to the same filtering process that was used to identify secreted proteins in the mass spectrometry data to generate a list of all proteins that are potentially secreted from any cell in the mouse. Fisher's exact test was used to identify overrepresented gene ontology terms in the identified proteins compared with this subset of the database. Molecular function terms occurring in the identified proteins at least 10 times and biological process terms occurring at least 20 times were tested at a significance level of 0.05 after correction for multiple testing using the Benjamini and Hochberg (78) method. To account for biases that may be introduced through the MS measurements, we performed identical bioinformatics analyses testing for overrepresented GO terms within the cellular (non-secreted) proteins identified in our screen. These proteins were tested using the same criteria and background database. Five categories, namely nucleotide binding, protein binding, ATP binding, cellular metabolic process, and primary metabolic process, were found significantly overrepresented in both analyses and were therefore subtracted from the lists of enriched GO terms within the secreted protein data set.
Unsupervised clustering was performed using the fuzzy c-means algorithm as implemented in the Mfuzz package (19) for R (20). Only regulated protein identifications, as defined by a MaxQuant significance value <0.05 in at least one time point, were subjected to clustering into five clusters with a fuzzification parameter of 3. For characterization of proteins in each cluster, “biological process” and “molecular function” GO terms were counted, and terms occurring only once in a cluster were omitted from the analysis.
From all identified hydroxyproline sites, high confidence sites were extracted by requiring a Mascot score >30 and a MaxQuant localization probability >0.75 (21). High confidence sites were partitioned into sites matching the proteotypic collagen hydroxyproline motif (XPG or GP), sites on fibrinogen-1, and a final group containing the remaining sites. WebLogo (22) was used to illustrate the amino acid distribution in the proximity of the hydroxyprolines in the three groups. SILAC ratios for hydroxyproline-containing peptides were normalized by the SILAC ratio for the parent proteins.
Immunostaining
Cells were seeded on coverslips in 6-well plates in DMEM deficient in Arg and Lys and supplemented with normal Arg (Arg0), Lys (Lys0), 10% dFBS, l-glutamine, and antibiotics. Differentiation was induced when reaching total confluence using 2% dFBS (23). On days 0, 2, and 5 of differentiation, cells were washed six times in serum-free DMEM and subsequently starved for 12 h in accordance with the treatment of cells for mass spectrometry analysis. At the end of starvation, cells were fixed for 10 min using 3.7% paraformaldehyde. Permeabilization and blocking were achieved using 1% Triton X-100 in PBS and 1% horse serum, 0.1% Tween 20 in PBS, respectively. The nucleic acid stain Sytox (Molecular Probes, Invitrogen) was used for nuclear visualization. Antibodies and dilutions used were myosin heavy chain (MHC) (H-300) (Santa Cruz Biotechnology sc-20641; 1:50), anti-troponin T (Sigma-Aldrich T6277; 1:50), and Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes, Invitrogen; 1:200). Images were obtained using a Zeiss LSM 510 Meta confocal laser scanning microscope.
RNA Purification, cDNA Synthesis, and Real Time Quantitative PCR (Q-PCR)
Total cellular RNA was purified using TRIzol (Invitrogen) according to the manufacturer's instructions. RNA was extracted from dishes of cells from which CM were collected for mass spectrometry analysis. 1 μg of total RNA was reversed transcribed using 200 units of Moloney MLV reverse transcriptase (Invitrogen) according to the supplier's protocol with 250 ng of random hexamer primers (GE Healthcare) but without any RNase inhibitor. Quantitative PCR analysis was performed using the SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) and run on the Mx3000P Q-PCR machine from Stratagene. The specificity and quality of primers were evaluated, including a dissociation curve analysis at the end of each PCR run. In addition, to ensure that primers only gave rise to a single product, amplicons were analyzed by ethidium bromide-stained 1.5% agarose gels. For normalization, the expression of the reference gene TATA box-binding protein (TBP) was used. Primers used for real time Q-PCR were as follows: IGF-1: forward primer, 5′-atg ctc ttc agt tcg tgt gtg g-3′; reverse primer, 5′-tct tgg gca tgt cag tgt gg-3′; IGF-2: forward primer, 5′-cta ctt cag cag gcc ttc aag c-3′; reverse primer, 5′-gga agt cgt ccg gaa gta cg-3′; TGF-β1: forward primer, 5′-ctg gat acc aac tat tgc ttc agc-3′; reverse primer, 5′-gca agg acc ttg ctg tac tgt g-3′; TGF-β2: forward primer, 5′-ctc ccc tcc gaa aat gcc-3′; reverse primer, 5′-gga tct gat aca gtt caa tcc gc-3′; TGF-β3: forward primer, 5′-gcg gag cac aat gaa ctg g-3′; reverse primer, 5′-tca tcc ggt cga agt atc tgg-3′; Sema3A: forward primer, 5′-cgg gaa cca aca act att tcg-3′; reverse primer, 5′-tgc gcc tct ttg cag tag g-3′; Sema3E: forward primer, 5′-gac ggc tac aga gag ata tac tgg c-3′; reverse primer, 5′-aac agg ggt tcc tct gaa tgg-3′; Sema6A: forward primer, 5′-tct acg ttg ctg ctc gag acc-3′; reverse primer, 5′-agg tat cga ccc tgt agt ttc tgc-3′; Sema7A: forward primer, 5′-ggc gga agc tct atg tga cc-3′; reverse primer, 5′-gac tgc agc act gat cgt tgg-3′; and TBP: forward primer, 5′-acc ctt cac caa tga ctc cta tg-3′; reverse primer, 5′-atg atg act gca gca aat cgc-3′.
Whole Cell Extracts and Western Blotting
Whole cell extracts were prepared using radioimmune precipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 0.25% sodium deoxycholate, 0.1% SDS, 1 mm Na3VO4, 5 mm β-glycerophosphate, 5 mm NaF) containing protease inhibitors (Roche Applied Science). Whole cell extracts were prepared from the same dishes of cells used for mass spectrometry studies. Approximately 25 μg of protein were separated using Novex 4–12% Bis-Tris gradient gels (Invitrogen). Antibodies and dilutions used were anti-troponin T (Sigma-Aldrich T6277; 1:1000), MHC (H-300) (Santa Cruz Biotechnology sc-20641; 1:1000), myogenin (F5D) (Santa Cruz Biotechnology sc-12732; 1:500), rabbit anti-semaphorin 3A (Millipore AB9604; 1:1000), anti-human semaphorin 3E (R&D Systems AF3239; 1:1000), rabbit polyclonal antibody to semaphorin 7A (Abcam ab23578; 1:2000), β-actin (C4) (Santa Cruz Biotechnology sc-47778; 1:5000), and HRP-conjugated secondary antibodies (GE Healthcare).
Conditioned Media for Western Blotting and Zymography
Cells were cultured as described above except that all cell populations (day 0, 2, and 5 cells) received normal Arg and Lys. Cells were washed six times and incubated in serum-free DMEM supplemented with penicillin/streptomycin and l-glutamine for 12 h on days 0, 2, and 5 during myogenesis. Differentiation was induced at confluence using 2% dFBS as for the proteomics experiments. CM were harvested centrifuged and filtrated (0.2-μm filter) to remove any dead cells and cell debris. Enrichment of secreted proteins was done via ultrafiltration using a 3000-Da molecular mass cutoff (Vivaspin columns, Sartorius). After concentration, the protein content of the individual media was estimated via Bradford assay (Coomassie Plus reagent, Pierce).
Western blot analysis of selected proteins was done by separating 25 μg of proteins via Novex 4–12% Bis-Tris gradient gels (Invitrogen). Equal loading of concentrated CM from days 0, 2, and 5 was verified by Colloidal Blue staining of a corresponding 4–12% Bis-Tris gradient gel. Antibodies and dilutions used were rabbit anti-semaphorin 3A (Millipore, AB9604; 1:2000), anti-human semaphorin 3E (R&D Systems AF3239; 1:1000), Sema6A (QQ06) (Santa Cruz Biotechnology sc-74274; 1:100), anti-human/mouse semaphorin 7A (CD108) (R&D Systems MAB2068; 1:1000), and HRP-conjugated secondary antibodies (GE healthcare).
In zymography studies, 5 μg of proteins were separated via 10% Tris-glycine gels containing 0.1% gelatin as substrate. CM were analyzed by one-dimensional electrophoresis without prior reduction of samples. Following gel electrophoresis, gels were washed using Zymogram Renaturing Buffer (Invitrogen) to remove SDS. Subsequently, gels were incubated in Zymogram Developing Buffer (Invitrogen) at 37 °C for approximately 12 h of digestion. Colloidal Blue staining (Invitrogen) was used to visualize the activity of proteases.
RESULTS AND DISCUSSION
Efficient Differentiation of C2C12 Cells in Labeling Media
Development of the skeletal muscle is a complex multistep process that is under the control of numerous extracellular stimuli (24, 25). Upon induction of differentiation, myoblasts withdraw from the cell cycle, differentiate, elongate, and fuse into multinucleated myotubes. Myogenesis requires the coordinated regulated expression of numerous genes and proteins. The myogenic program is orchestrated by a family of transcription factors known as myogenic regulatory factors (MRFs), including MyoD, Myf-5, myogenin, and MRF4/Myf-6 (26, 27). In association with other transcription factors such as the myocyte enhancer factor 2 (MEF2) family, MRFs modulate the transcriptional activity of many muscle-specific genes. In addition to MRFs, muscle-secreted proteins also play a pivotal role, affecting local muscle biology in an autocrine and paracrine manner (24, 28).
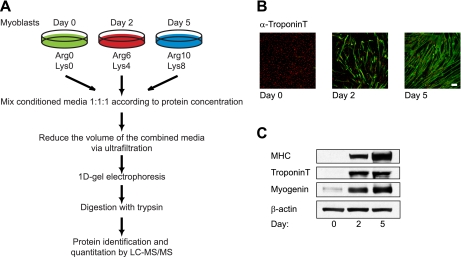
In this study, we utilized the triple encoding SILAC approach during muscle differentiation of murine C2C12 cells induced by using 2% dialyzed serum (Fig. 1A) (see “Experimental Procedures”). The efficiency of differentiation was verified by analyzing the expression of well known myogenic markers either via confocal laser scanning microscopy (Fig. 1B) or via Western blotting (Fig. 1C). Fusion of myoblasts into small myotubes was visible by day 2 of differentiation, and by day 5 of differentiation, the myoblasts were engaged in formation of long multinucleated myotubes (Fig. 1B). Additionally, expression of troponin T and MHC, two proteins involved in muscle physiology, and the muscle-regulatory transcription factor myogenin was enhanced on day 2 and throughout differentiation (Fig. 1C).
Fig. 1.
Myogenic commitment of C2C12 myoblasts using dFBS. A, experimental overview. C2C12 cells were grown to confluence (day 0), and myoblast differentiation was induced by reducing the amount of dFBS to 2%. CM were collected on days 0, 2, and 5 of differentiation of C2C12 cells. “Day 0 cells” were cultured using regular Arg0 and Lys0, “day 2 cells” were cultured using Arg6 and Lys4, and “day 5 cells” were cultured using Arg10 and Lys8. CM were concentrated by ultrafiltration, and proteins were separated by one-dimensional gel electrophoresis. Excised gel bands from the entire lane were subjected to in gel-digestion and subsequently analyzed on an LTQ-Orbitrap-XL mass spectrometer. B and C, efficiency of differentiation was verified by examining the expression of muscle markers either by confocal laser scanning microscopy (B) or Western blotting (C). B, immunostaining of C2C12 cells at days 0, 2, and 5 of differentiation with an antibody against troponin T and visualized by a green fluorescence-tagged secondary antibody. Nuclei were counterstained with Sytox. The scale bar represents 95 μm. C, Western blot analysis of whole cell extracts prepared from C2C12 myoblasts at days 0, 2, and 5 of myogenesis using the indicated antibodies.
Identification of Murine Skeletal Muscle Secretome
We applied triple encoding SILAC to murine C2C12 cells during the course of myogenesis to investigate which proteins are secreted and regulated during skeletal muscle differentiation (Fig. 1A). One of the major challenges in secretome studies is to distinguish cell-derived proteins from residual serum proteins. Applying the SILAC approach offers an advantage compared with other labeling strategies because metabolic labeling of cells facilitates the discrimination between secreted proteins versus contaminating serum proteins due to the incorporation of the isotope-labeled amino acids into the proteins during their synthesis inside the cell. To quantitatively analyze protein secretion during myogenesis, C2C12 cells were cultured in Arg- and Lys-deficient medium supplemented with either normal Arg0 and Lys0 (CM collected at day 0) or isotope-labeled analogues Arg6 and Lys4 (CM collected at day 2 of differentiation) and Arg10 and Lys8 (CM collected at day 5 of differentiation). Using high resolution mass spectrometry, the MaxQuant software suite for data processing and quantitation, and the ProteinCenter bioinformatics program for protein annotations, we identified 635 putative secreted proteins (supplemental Table 1). The list of identified secreted proteins from skeletal myoblasts in this study represents the largest number of muscle-secreted proteins reported so far. In addition, we were able to quantitatively profile 624 of these proteins, thereby revealing the dynamics of their secretion during the course of myogenesis (supplemental Table 1).
Functional Annotation Analyses of Identified Secreted Proteins
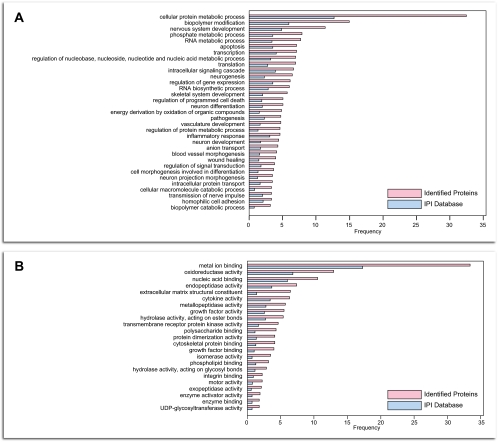
We next characterized the distribution of the skeletal muscle secretome according to the GO biological process and molecular function annotations and performed statistical enrichment analyses to identify overrepresented categories in our data set. We first extracted all potential secreted proteins from the entire IPI mouse database applying the same criteria for selecting secreted proteins as for the proteins identified in this work (see “Experimental Procedures”) and used these as a reference data set.
The identified secreted proteins were found to be functionally diverse and to be involved in various biological processes. The significant enrichment of GO categories like regulation of signal transduction (n = 24, p = 0.001), inflammatory response (n = 29, p = 0.04), skeletal system development (n = 36, p = 3.2e−7), and neurogenesis (n = 41, p = 2.8e−8) highlights distinct roles of the secreted muscle proteins (Fig. 2A and supplemental Table 2). Moreover, several molecular function categories were found to be significantly enriched compared with the IPI database, further demonstrating an enrichment of distinct proteins with a defined function in our data set (Fig. 2B and supplemental Table 2). Not surprisingly, among the most overrepresented terms were the metal ion binding (n = 212, p = 9.5e−10), the extracellular matrix structural constituent (n = 41, p = 1.1e−12), and the enzyme regulator activity (n = 54, p = 1.4e−7). Key categories, including enzyme inhibitor activity (n = 40, p = 6.4e−6), enzyme activator activity (n = 12, p = 0.01), integrin binding (n = 14, p = 0.01), growth factor activity (n = 35, p = 0.002), growth factor binding (n = 25, p = 1.4e−6), cytokine activity (n = 40, p = 0.01), and metallopeptidase activity (n = 36, p = 0.003), were also found to be significantly overrepresented. These findings clearly characterize the skeletal muscle as an important secretory organ in the body. For example, we found several pivotal proteins with growth factor activity such as activin, transforming growth factor (TGF)-β, FGF-21, macrophage colony-stimulating factor, and platelet-derived growth factor (PDGF).
Fig. 2.
Functional analyses of identified secreted proteins from C2C12 skeletal myoblasts. A and B, identified secreted proteins from C2C12 myoblasts were compared with a reference list of secreted proteins obtained from the entire list of IPI entries. For selected biological processes (A) and molecular functions (B), the frequency of the terms in the data and the IPI database is illustrated. Only significantly overrepresented categories (p < 0.05) are shown.
Characterization of Differentially Regulated Secreted Proteins during Myogenesis
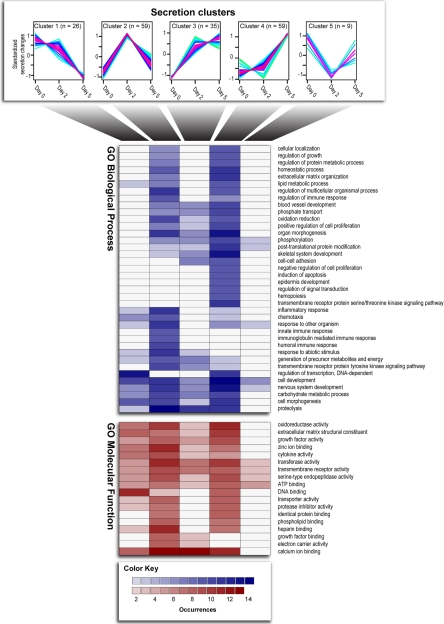
Statistical analysis revealed 188 of the 624 secreted proteins to be differentially regulated between proliferating and differentiating myoblasts (Fig. 3 and supplemental Table 3). Regulated proteins were subjected to unsupervised clustering according to their relative secretion profile, resulting in five distinct dynamic profiles. In general, a higher proportion of the identified muscle-released proteins exhibited an increased level of secretion compared with the proteins with a decreased secretion profile during the course of C2C12 differentiation (Fig. 3). More than 60% of the proteins were assigned to two clusters: cluster 2 demonstrates a marked increase in secretion during early differentiation, and cluster 4 represents secreted proteins with a pronounced up-regulation in secretion levels at terminal differentiation. This observation indicates that an overall increased secretory activity is accompanying skeletal muscle cell maturation.
Fig. 3.
Dynamic and functional characterization of differentially regulated secreted proteins. Cluster analysis of regulated proteins according to their secretion profile is shown. The heat map summarizes the occurrences of the indicated biological process and molecular function GO terms within each cluster. Only GO terms with five or more occurrences in at least one cluster are shown.
To examine whether proteins with similar biological implications also demonstrated similar regulation, we looked at the distribution of the regulated secreted proteins into the GO annotations biological processes and molecular function (Fig. 3 and supplemental Table 3). Upon induction of differentiation, a majority of the C2C12 myoblasts withdraw from the cell cycle and fuse to form long multinucleated myotubes. In agreement with this, we identified several secreted proteins classified as negative regulators of cell proliferation to be up-regulated at terminal differentiation. In contrast, we found that secreted proteins involved in the positive regulation of proliferation exhibited a more diverse profile, allocating to three different clusters demonstrating both a reduced secretion and an increased secretion during the course of differentiation, illustrating the complexity of the myogenic program. The formation of long multinucleated myotubes during muscle differentiation is a critical process that involves multiple steps such as cell migration, alignment, recognition, adhesion, cell fusion, and reorganization of the ECM. We observed several proteins involved in cell-cell adhesion and extracellular matrix organization exhibiting differential secretion profiles during the course of differentiation. In addition, we also found differential secretion of several proteins annotated to play a role in different metabolic processes, organ morphogenesis, cell development, cell morphogenesis, blood vessel development, and nervous system development, all biological processes connected with cell differentiation and tissue development (Fig. 3).
Differential Secretion of Myogenic Regulatory Growth Factors during Myoblast Differentiation
Skeletal myogenesis is modulated by a number of growth factors, which may act in an autocrine and/or paracrine manner to regulate proliferation and differentiation. TGF-β and insulin-like growth factor (IGF) are two groups of growth factors demonstrated to play critical regulatory roles in the control of muscle development either inhibiting or promoting differentiation, respectively.
Dynamic Secretion Profile of TGF-β1, -β2, -β3, and Associated Regulatory Proteins
TGF-βs are small 25-kDa pleiotropic proteins involved in a wide array of physiological processes, including cell proliferation, differentiation, angiogenesis, migration, and apoptosis (29, 30). In addition, TGF-β also plays an important role in ECM morphogenesis, promoting the synthesis of ECM components. TGF-β can function in an autocrine and paracrine way as well as targeting more distant cells.
We identified all three isoforms of TGF-β (TGF-β1, -β2, and -β3) as being secreted and differentially regulated during myogenic conversion (Fig. 4A and supplemental Table 1). Little is known concerning the role of the individual isoforms of TGF-β in myogenesis, although all three isoforms of TGF-β have been shown to function as inhibitors of skeletal muscle differentiation in vitro (31–33). Only under certain specialized conditions, including serum-rich media or low cell density, was TGF-β found to enhance differentiation (34, 35). Recently, in vivo studies also supported TGF-β as a negative regulator of muscle development during muscle regeneration (36).
Fig. 4.
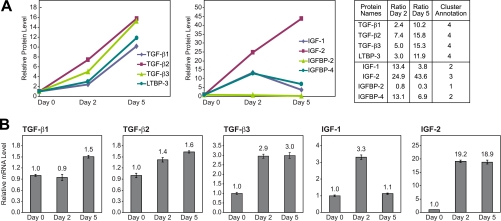
Secretion and expression of TGF-β1, -β2, and -β3 and IGF-1 and -2 are differentially regulated during the course of skeletal muscle differentiation. A, dynamic secretion profiles, including ratios obtained by mass spectrometry analyses, for the proteins TGF-β1, -β2, -β3, LTBP-3, IGF-1, IGF-2, IGFBP-2, and IGFBP-4 during the course of C2C12 differentiation. B, mRNA expression of TGF-β and IGF isoforms at days 0, 2, and 5 of C2C12 differentiation analyzed by Q-PCR. The level of mRNA expression of selected genes was normalized to the mRNA level of TBP. Gene expression data are given relative to day 0, and data are shown as mean ± S.D. (n = 3). Error bars represent the Standard Deviation S.D.
Interestingly, we observed a pronounced increase in the secreted level of TGF-β1, -β2, and -β3 during myogenesis (Fig. 4A). This apparent paradox clearly illustrates the complexity of TGF-β signaling in the myogenic program. The secretion of TGF-β2 and -β3 increased gradually during the course of differentiation, whereas TGF-β1 secretion was similar on days 0 and 2 of differentiation but increased 10-fold on day 5. Other studies have also demonstrated an increased expression of TGF-β in skeletal muscle during myogenesis and regeneration (31, 37–39). It should be emphasized, however, that an increased level of secreted TGF-β does not necessarily reflect an increased level of bioactive TGF-β. The level of TGF-β signaling is regulated in many different ways, one of them being sequestration of TGF-β to the ECM via latent TGF-β-binding proteins (LTBPs). We found all four LTBP members to be secreted from the C2C12 cells. Although the secretion of LTBP-1, LTBP-2, and LTBP-4 was not significantly regulated during myogenesis, the release of LTBP-3 was modestly enhanced on day 2 but markedly increased on day 5 (Fig. 4A and supplemental Table 1). The function of the individual LTBPs in skeletal myogenesis still needs to be elucidated, and it is tempting to speculate that they have a major role in stabilizing the muscle-released TGF-β for endocrine purposes toward more distant target tissues.
In addition to the family of LTBP proteins, we also identified other proteins known to function as modulators of TGF-β activity exhibiting a differential secretion profile during differentiation. Decorin and biglycan are two small proteoglycans residing in the ECM and reduce the bioavailability of TGF-β during myogenesis (40, 41). We found that the level of secreted decorin was markedly increased during the early stages of differentiation, whereas an increased secretion of biglycan was observed throughout differentiation (supplemental Table 1).
Dynamic Secretion Profile of IGF-1 and IGF-2
In contrast to the TGF-βs, IGFs known to promote differentiation demonstrated an increased level of secretion in agreement with the functionality of IGF in myogenesis. The growth factors IGF-1 and IGF-2 are known to be secreted from muscle cells and are both involved in numerous processes in skeletal muscle biology (42, 43). In addition, recent studies have suggested that IGF-1 may play a more significant role in metabolic homeostasis, including skeletal muscle metabolism, than previously anticipated (44, 45). Here we show the dynamic profile of IGF secretion during a 5-day differentiation study of C2C12 myoblasts (Fig. 4A and supplemental Table 1). Secretion of IGF-2 increased dramatically during the course of differentiation, supporting previous findings that IGF-2 plays an essential regulatory role throughout differentiation. In contrast, the highest level of secreted IGF-1 was observed on day 2 of differentiation, indicating that IGF-1 is a more important modulator of cellular processes in the initial phases of differentiation.
In the extracellular environment, the activity of IGFs is regulated by IGF-binding proteins (IGFBPs), which modulate the bioactivity of IGFs (46, 47). The IGFBPs exert a dual function by either inhibiting or enhancing the action of IGFs depending on cell type and context. Interestingly, several studies have demonstrated that IGFBPs also exert IGF-independent cellular actions modulating gene expression and activating intracellular signaling. We identified five IGFBPs (IGFBP-2, IGFBP-4, IGFBP-5, IGFBP-6, and IGFBP-7) as proteins secreted from C2C12 cells; the secretion of IGFBP-2 and IGFBP-4 was found to be differentially regulated during the course of differentiation (Fig. 4A and supplemental Table 1). Increased amounts of secreted IGFBP-4 were observed on day 2 of differentiation followed by a reduction on day 5 but still 6-fold higher compared with day 0 (Fig. 4A). IGFBP-2 has in many studies been shown to serve a growth-promoting function (48). In support of this, we found that the level of secreted IGFBP-2 indeed decreased during the course of differentiation in which cells withdraw from the proliferative phase (Fig. 4A).
The differential secretion pattern of the IGFBPs suggests that they serve distinct functions in muscle development. Expression of IGFBP-5 increases dramatically during differentiation, and IGFBP-5 has attracted a lot of attention in a number of studies investigating muscle physiology where it was shown to be either a negative or positive regulator of differentiation depending on the experimental conditions (49–52). However, we did not observe any significant regulation of the released IGFBP-5 during differentiation (supplemental Table 1). In addition, we identified IGFBP-6 and IGFBP-7 to be secreted from skeletal myoblasts, but we did not observe any significant regulation of their secretion during the process of differentiation (supplemental Table 1).
Secretion of IGFs and TGFs Is Regulated by Posttranscriptional Mechanisms
Several studies have shown that the dynamic profile of protein expression often cannot be correlated to dynamics at the RNA level (53, 54). Therefore, we decided to investigate whether the increased release of IGF and TGF-β was regulated at the mRNA level (Fig. 4B). Examining the expression of TGF-β mRNA using Q-PCR revealed a very modest up-regulation of TGF-β1 expression on day 5 and of TGF-β2 on both days 2 and 5. TGF-β3 demonstrated a more pronounced increase in expression although not to the same extent as the level of secreted TGF-β3. In agreement with previous reports, we found a dramatic up-regulation of IGF-2 mRNA during myogenesis (52, 55). Whereas the mRNA expression of IGF-2 was similar on both days 2 and 5 of differentiation, we observed a higher level of secreted IGF-2 on day 5 compared with day 2. Moreover, secretion of IGF-2 was markedly higher compared with RNA expression at terminal differentiation. Although the dynamics of IGF-1 secretion resembles its mRNA levels, the secreted IGF-1 was further increased severalfold on day 2 compared with the corresponding increase of mRNA expression (Fig. 4). Collectively, these data suggest that the differential release of IGFs and TGF-βs is predominantly regulated by posttranscriptional mechanisms.
Secretion of Semaphorins during Muscle Development
The mammalian family of semaphorins is a large group of proteins consisting of five subclasses, including secreted (Sema3A–G), transmembrane (Sema4A–G, Sema5Aand Sema5B, and Sema6A–D), and glycosylphosphatidylinositol-anchored (Sema7A) proteins. The semaphorins were initially characterized as constituents of the regulatory system responsible for axon guidance during the development of the nervous system (56). However, accumulating data have now recognized semaphorins as important modulators of angiogenesis, organogenesis, tumor progression, and immune responses (57). Moreover, recent studies have suggested that members of the semaphorin family play an important role in regulating skeletal muscle development (58, 59).
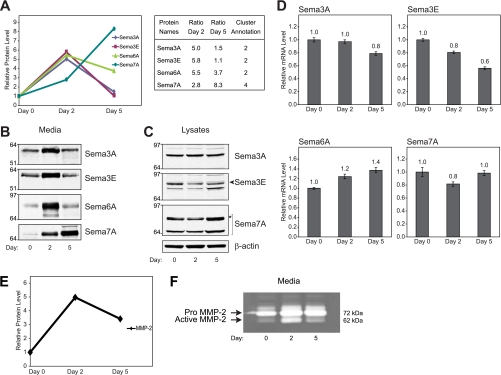
We identified four members of the Sema3 subfamily (Sema3A, Sema3B, Sema3D, and Sema3E), two members of the Sema4 subfamily (Sema4B and Sema4C), and one member of each of the Sema6 and Sema7 subfamilies (Sema6A and Sema7A) to be secreted from both myoblast and myotubes (Fig. 5A and supplemental Table 1). Moreover, the secretion of the soluble Sema3A, Sema3D, and Sema3E and membrane-associated Sema6A and Sema7A was found to be differentially regulated during the course of differentiation. The secretion of Sema3A, Sema3E, Sema3D, and Sema6A was markedly elevated on day 2 of differentiation, whereas the secretion of Sema7A increased throughout differentiation (Fig. 5A). Western blot analyses examining the level of secreted Sema3A, -3E, -6A, and -7A demonstrated the same trend of dynamics as the dynamic profiles obtained by mass spectrometry (Fig. 5B). Interestingly, analyzing the intracellular protein expression of Sema3A, -3E, and -7A demonstrated a somewhat different profile, suggesting that the level of secreted semaphorin is regulated posttranslationally (Fig. 5C). Although protein expression of Sema3A remained more or less constant during myogenesis, the expression of Sema3E was decreased at both days 2 and 5 of differentiation. The expression of Sema7A displayed a similar profile to that of secreted Sema7A, although the increase in secreted Sema7A was more pronounced compared with the increase in protein expression.
Fig. 5.
Differential secretion of different subclasses of semaphorin proteins during the process of myogenesis. A, quantitative profiles of semaphorin secretion during the course of C2C12 differentiation, including their ratios, applying quantitative proteomics. B and C, Western blot analysis of selected semaphorin proteins in conditioned media (B) or whole cell extracts (C) at days 0, 2, and 5 of C2C12 differentiation. D, mRNA expression for Sema3A, Sema3E, Sema6A, and Sema7A by real time Q-PCR. Gene expression data were normalized to the expression of TBP and are presented as mean ± S.D. (n = 3) relative to day 0. E, dynamic profile of MMP-2 secretion as observed by MS. F, gelatin zymography assessing MMP-2 activity in conditioned media from C2C12 myoblasts at days 0, 2, and 5 of differentiation (pro-MMP-2, 72 kDa; and active MMP-2, 62 kDa). * represents the glycosylated form of Sema7A.
Investigating the mRNA expression of Sema3A, Sema3E, and Sema7A by Q-PCR indicated that the level of secreted semaphorins is regulated both by posttranscriptional as well as posttranslational mechanisms (Fig. 5D). The expression profiles of Sema3A and Sema3E comparing RNA and protein were similar, implying that the high level of secreted Sema3A and Sema3E on day 2 of differentiation occurs via posttranslational regulatory mechanisms (Fig. 5, A–D). The expression of Sema7A mRNA remained constant during differentiation, whereas the expression of Sema7A protein was increased at day 5 of differentiation. In addition, the level of secreted Sema7A was increased at day 2 and further increased at day 5, indicating that secreted Sema7A is regulated both at the posttranscriptional and posttranslational levels (Fig. 5, A–D). Collectively, these data demonstrate that care should be taken trying to extrapolate the level of secreted proteins from intracellular protein expression. The differences observed between intracellular protein expression and the secreted proteins indicate that the level of individual secreted proteins can be regulated either via the secretory pathway or by stability and/or processing of released proteins.
The high level of secreted Sema3A, Sema3E, and Sema6A observed on day 2 suggests that they may function in an autocrine fashion regulating early events during differentiation, whereas the increased secretion of Sema7A throughout differentiation indicates that Sema7A may play a role during early and late events of myogenesis. Recent studies have shown that Sema7A is involved in bone morphogenesis promoting migration of osteoblasts and enhancing fusion of chondrocytes during differentiation, two cellular processes that are required for skeletal muscle differentiation (60). Interestingly, the increased release of semaphorins could also serve an important paracrine function (59, 61). Several studies have shown that semaphorins are important regulators of cell migration, neurogenesis, and angiogenesis, processes essential for skeletal muscle development and regeneration (57, 62). In a recent work by Tatsumi et al. (59), it was speculated that the increased expression of Sema3A observed in satellite cells in response to injury-induced muscle regeneration could serve a paracrine function regulating muscle innervation. Moreover, Sema6C was found to localize primarily at neuromuscular junctions in vivo, and the expression of Sema6C was reduced in response to denervation of skeletal muscle, indicating that other classes of semaphorins could be involved in neurogenesis of skeletal muscle (63).
Class 3 Semaphorins Are Involved in Activation of Matrix Metalloproteinase (MMP)-2 Activity in Skeletal Muscle
Studies have shown that proteolytic processing of class 3 semaphorins modulates their activity (64, 65). In turn, class 3 semaphorins themselves transmit some of their effects by activating a specific group of proteases, the MMPs (66, 67). The growth-promoting effect of Sema3A in cortical dendrites relates to activation of MMP-2. The family of MMPs, including MMP-2, is involved in ECM morphogenesis, release of bioactive signaling molecules from the ECM, shedding of membrane-associated proteins, and cell motility, all indispensable processes for the integrity of muscle development and regeneration (68, 69). Secretion of MMP-2 was increased during myogenesis, demonstrating the highest level during early differentiation (Fig. 5E). To determine whether the increased secretion of MMP-2 correlated with increased activity of MMP-2, we carried out gelatin zymography analyzing conditioned media collected at days 0, 2, and 5 of differentiation (Fig. 5F). The zymograms revealed that the increased level of released MMP-2 was associated with increased protease activity of MMP-2, which reached its maximum at day 2 of differentiation. It has been demonstrated that Sema3A enhances growth of cortical dendrites via the induction of both protein expression and activity of MMP-2 (67). In contrast, Sema3A decreased both the expression and activity of MMP-3, whereas Sema3C promoted axonal growth by activating MMP-3 (66, 67). Comparing the dynamic profile of Sema3A release with the profiles of MMP-2 secretion and activity suggests that the activation of MMP-2 by Sema3A signaling might occur in cell types other than dendrites, including skeletal muscle cells. Moreover, other members of the identified class 3 semaphorins could be involved in modulating the activity and response to metalloproteases. Additionally, MMPs can also function as regulators of semaphorin signaling cascades by cleavage of semaphorin proteins. Soluble Sema4D promotes angiogenesis after being shed from the membrane, a process involving MMP-14 activity (70, 71), whereas Sema7A was identified as a substrate of MMP-2 (72).
Identification of Hydroxyproline Sites on Proteins Secreted by Myoblasts and Myotubes
Regulation of protein activity by posttranslational modifications such as phosphorylation and ubiquitination is well known. However, recent findings indicate that proline hydroxylation also serves an important regulatory role modulating protein activity exemplified by the transcription factor hypoxia-inducible factor (73). The stability of the oxygen-regulated subunit hypoxia-inducible factor α is modulated by hydroxylation of a conserved proline residue in response to the oxygen level within the cells.
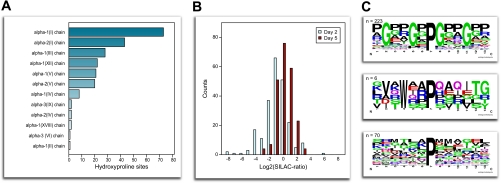
We identified a total of 299 unique high confidence hydroxyproline sites from 48 distinct secreted proteins (supplemental Table 4 and annotated hydroxyproline MS/MS spectra). Notably, 231 of these sites were derived from various collagen types with a large variation of the number of modified sites per individual protein, ranging from one on collagen α-3 type VI to over 70 on collagen α-1 type I (Fig. 6A). Collagens are the most abundant constituents of the ECM, and the collagen triple helix structure consists of three polyproline chains comprising the repetitive sequence motif XYG where the X and Y positions are frequently occupied by proline and its posttranslational modification 4-hydroxyproline, respectively. The presence of hydroxyproline residues in collagens has been known for decades, and both proline and hydroxyproline are essential for formation and stabilization of the collagen triple helix structure (74). Interestingly, we found an overall down-regulation in oxidation of proline residues on day 2 of C2C12 differentiation compared with days 0 and 5 (p = 3e−14) (Fig. 6B). This could reflect an increased level of newly synthesized proteins, which have not yet been posttranslationally modified, and the formation of less rigid ECM structures to allow for the early events during fusion and formation of shorter myotubes. At terminal differentiation in which the majority of myoblasts are engaged in the formation of long multinucleated myotubes, the reorganization and stabilization of the ECM are more defined. Accordingly, we observed an overall increase of the hydroxyproline residues on collagen molecules on day 5 of differentiation.
Fig. 6.
Analyses of hydroxyproline sites identified from proteins secreted by C2C12 myoblasts and myotubes. A, distribution of hydroxyproline sites on the different collagen subtypes. B, distribution of the regulation (SILAC ratios) of hydroxyproline sites at days 2 (mean, −1.49) and 5 (mean, −0.32) of differentiation. C, hydroxyproline motif analysis. Sites matching the prototypical collagen hydroxyproline motif (top panel), sites with a tryptophan in the −3 position, all on fibronectin (middle panel), and the remaining sites not matching a significant shared motif (bottom panel) are shown.
We next performed motif sequence analysis to identify specific linear hydroxyproline sequence motifs. Alignment of the sequences surrounding the identified 299 hydroxyproline sites exposed a motif corresponding to the canonical motif previously reported for collagen proteins (Fig. 6C, top panel). The presence of the tripeptide sequence XPG (n = 223) was identified exclusively on collagens (Fig. 6C). In addition, we identified a novel hydroxyproline motif containing a tryptophan residue on the third position preceding the modified proline (Fig. 6C, middle panel). The tryptophan at the −3 position was found only on peptides from fibronectin, another major compound of the ECM. Analysis of the remaining 70 hydroxyproline sites did not reveal additional sequence motifs (Fig. 6C, bottom panel). These sites spread over more than 40 proteins, including fatty acid-binding protein; several components of the ECM such as SPARC, fibronectin, Lama2, and perlecan; and different inhibitors of proteolytic enzymes such as serine protease inhibitors (serpinf1 and serphinh1) and the metalloproteinase inhibitor 2 (Timp2). The two identified hydroxyproline motifs were found only on collagens and fibronectin, respectively, both components of the extracellular matrix. This could indicate no enzyme recognition motif requirements as it may arise merely from the need for repetitive sequences for proper function of the ECM components. In addition, the majority of the identified hydroxyproline sites are not known as evident from the UniProt database, and although beyond the scope of this study, it will be interesting to assess the role of these posttranslational modifications for the function of the corresponding secreted proteins.
Conclusions
Proteins secreted from skeletal muscle influence local muscle biology and have a systemic effect on other organs, including brain, adipose tissue, and liver. The cross-talk between tissues occurs via signaling molecules that coordinate the major cellular processes such as cell growth, differentiation, and survival. The dynamic orchestra of events underlying the proper communication between cells and tissues depends entirely on the release of secreted factors, which consequently trigger appropriate cellular responses in the target cells. The fat-muscle-liver-brain tissue cross-talk is of particular interest as it can shed light on the missing links in the development of a number of metabolic disorders and complications. Since the discovery that the adipose tissue is an endocrine organ, tremendous focus has been drawn to the secretory function of adipocytes and the role of adipokines in obesity, diabetes, and inflammation (75, 76). Although the muscle represents the largest organ in the body, its role as an active endocrine organ that secretes factors with an auto-, para- and/or endocrine function has been recently recognized with the discovery of IL-6 (77). We applied the quantitative proteomics platform SILAC to analyze the muscle secretome, resulting in identification of 635 secreted proteins from muscle cells. Not surprisingly, the diversity of identified proteins reflects well the plethora of effects they exert and the various processes they control. The identified factors have distinct molecular functions, including growth factor activity, cytokine activity, enzyme activity, and components of the ECM, and are involved in the regulation of different cellular processes, ranging from angiogenesis to neurogenesis and the inflammatory response. The overrepresentation of specific molecular functions and biological processes in our data set underlines the significance of the skeletal muscle as a prominent secretory organ.
The dynamic secretion profile shown for 624 of the secreted factors provides valuable information for the changes of the protein expression and secretion during myoblast conversion. We compared the levels of the secreted factors at initial, medium, and terminal stages of myogenesis and found 188 proteins to be significantly regulated during the course of differentiation. The regulated proteins clustered into five specific groups representing the dynamic changes occurring in the secretome. The mass spectrometry-driven platform allowed the characterization of entire families of proteins as well as their regulatory partners. The identification of all three isoforms TGF-β1, -β2, and -β3 emphasizes another major advantage of our mass spectrometry-based proteomics analyses: the possibility to identify individual isoforms of the same protein based on identification of unique peptides found in each isoform. The number of commercially available antibodies keeps rising; however, there is no guarantee for successful functionality of these antibodies. In addition, generating antibodies recognizing specific isoforms can prove to be quite difficult and labor-intensive, especially if large homology exists between protein isoforms. Verifying whether an antibody targets a specific isoform or recognizes all isoforms is difficult to distinguish by Western blotting if the molecular weight of protein isoforms only differs slightly. Therefore, the mass spectrometry-based proteomics is the method of choice when investigating the secreted protein content.
We followed the dynamics of the TGF-β superfamily members and their modulators such as LTBPs, decorin, and biglycan as well as IGF family members and their regulators, the IGFBPs. Investigation of the expression profiles of specific secreted proteins demonstrated that the level of secreted proteins is regulated posttranscriptionally at several levels. We showed several examples of mRNA expression patterns that did not correlate to their corresponding secretion profile, indicating regulation by posttranscriptional mechanisms. Furthermore, analyzing the intracellular levels of semaphorin protein expression and comparing them with the dynamics of their secretion clearly illustrates that the release of secreted proteins is subjected to complex modular regulation.
One of the major strengths of mass spectrometry-based proteomics is the possibility to identify various posttranslational modifications. We identified 299 hydroxyproline sites; the majority of those were found on different collagen subtypes. However, we also identified hydroxyproline sites on many other proteins, including several constituents and regulatory components of the ECM, which could serve as important modifications to confer protein stability and interaction with other proteins. The identification of hydroxyproline on such a large number of peptides could be an indication for a more important regulatory role of this type of modification than previously anticipated.
The large number of identified secreted proteins in this study provides a foundation for future research to understand the regulation of the myogenic program. Understanding the role played by individual secreted proteins in skeletal muscle development could offer valuable clues in the search for new target proteins with therapeutic potential for treatment of various muscular disorders, including muscular dystrophies characterized by muscle atrophy and impaired regeneration. In addition, the secreted skeletal muscle proteins could also provide valuable candidates in the search for novel proteins exhibiting a systemic effect modulating the activity of other organs. It is anticipated that the identification of secreted proteins from skeletal muscle cells could allow greater insight into development of therapeutic strategies for patients with metabolic and muscle-related pathologies.
Supplementary Material
Acknowledgments
We thank all members from The Centre of Inflammation and Metabolism and the Center for Experimental BioInformatics for useful discussions. We are grateful to Dr. Carmen de Hoog (University of British Columbia, Vancouver, British Columbia, Canada), Dr. Jesper Olsen (The Novo Nordisk Foundation Center, Copenhagen, Denmark), and Dr. Joern Dengjel (Freiburg Institute for Advanced Studies, Freiburg, Germany) for critical reading of the manuscript. The Centre of Inflammation and Metabolism is supported by a grant from the Danish National Research Foundation.
Footnotes
* This work was supported by a grant from the Lundbeck Foundation, the Novo Nordisk Foundation, the Danish Natural Science Research Council, and the Danish Medical Research Council.
 This article contains supplemental Tables 1–4 and annotated hydroxyproline MS/MS spectra.
This article contains supplemental Tables 1–4 and annotated hydroxyproline MS/MS spectra.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- CM
- conditioned media
- dFBS
- dialyzed FBS
- ECM
- extracellular matrix
- GO
- gene ontology
- IGF
- insulin-like growth factor
- IGFBP
- IGF-binding protein
- IGF-1R
- IGF-1 receptor
- IPI
- International Protein Index
- LTBP
- latent TGF-β-binding protein
- LTQ
- linear trap quadrupole
- MMP
- matrix metalloproteinase
- MRF
- myogenic regulatory factor
- Q-PCR
- quantitative PCR
- Arg6
- l-[13C6,14N4]arginine
- Arg10
- l-[13C6,15N4]arginine
- Lys4
- l-[2H4]lysine
- Lys8
- l-[13C6,15N2]lysine
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TBP
- TATA box-binding protein
- Sema
- semaphorin.
REFERENCES
- 1.Pedersen B. K. (2009) The diseasome of physical inactivity—and the role of myokines in muscle-fat cross talk. J. Physiol. 587, 5559–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steensberg A., van Hall G., Osada T., Sacchetti M., Saltin B., Klarlund Pedersen B. (2000) Production of interleukin-6 in contracting human skeletal muscles can account for the exercise-induced increase in plasma interleukin-6. J. Physiol. 529, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen B. K., Febbraio M. A. (2008) Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol. Rev. 88, 1379–1406 [DOI] [PubMed] [Google Scholar]
- 4.Hojman P., Pedersen M., Nielsen A. R., Krogh-Madsen R., Yfanti C., Akerstrom T., Nielsen S., Pedersen B. K. (2009) Fibroblast growth factor-21 is induced in human skeletal muscles by hyperinsulinemia. Diabetes 58, 2797–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh K. (2009) Adipokines, myokines and cardiovascular disease. Circ. J. 73, 13–18 [DOI] [PubMed] [Google Scholar]
- 6.Chan X. C., McDermott J. C., Siu K. W. (2007) Identification of secreted proteins during skeletal muscle development. J. Proteome Res. 6, 698–710 [DOI] [PubMed] [Google Scholar]
- 7.Hittel D. S., Berggren J. R., Shearer J., Boyle K., Houmard J. A. (2009) Increased secretion and expression of myostatin in skeletal muscle from extremely obese women. Diabetes 58, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon J. H., Yea K., Kim J., Choi Y. S., Park S., Lee H., Lee C. S., Suh P. G., Ryu S. H. (2009) Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics 9, 51–60 [DOI] [PubMed] [Google Scholar]
- 9.Dengjel J., Kratchmarova I., Blagoev B. (2009) Receptor tyrosine kinase signaling: a view from quantitative proteomics. Mol. Biosyst. 5, 1112–1121 [DOI] [PubMed] [Google Scholar]
- 10.Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 11.Krüger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., Mann M. (2008) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokhorova T. A., Rigbolt K. T., Johansen P. T., Henningsen J., Kratchmarova I., Kassem M., Blagoev B. (2009) Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol. Cell. Proteomics 8, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blagoev B., Mann M. (2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243–250 [DOI] [PubMed] [Google Scholar]
- 14.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 15.Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 16.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 17.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 18.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar L., Futschik M. E. (2007) Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team (2007) R: a Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 21.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 22.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24.Chargé S. B., Rudnicki M. A. (2004) Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 25.Jansen K. M., Pavlath G. K. (2008) Molecular control of mammalian myoblast fusion. Methods Mol. Biol. 475, 115–133 [DOI] [PubMed] [Google Scholar]
- 26.Berkes C. A., Tapscott S. J. (2005) MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16, 585–595 [DOI] [PubMed] [Google Scholar]
- 27.Tapscott S. J. (2005) The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 28.Hawke T. J., Garry D. J. (2001) Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 91, 534–551 [DOI] [PubMed] [Google Scholar]
- 29.Kollias H. D., McDermott J. C. (2008) Transforming growth factor-beta and myostatin signaling in skeletal muscle. J. Appl. Physiol. 104, 579–587 [DOI] [PubMed] [Google Scholar]
- 30.Massagué J. (2008) TGFbeta in cancer. Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafyatis R., Lechleider R., Roberts A. B., Sporn M. B. (1991) Secretion and transcriptional regulation of transforming growth factor-beta 3 during myogenesis. Mol. Cell. Biol. 11, 3795–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart J. D., Masi T. L., Cumming A. E., Molnar G. M., Wentworth B. M., Sampath K., McPherson J. M., Yaeger P. C. (2003) Characterization of proliferating human skeletal muscle-derived cells in vitro: differential modulation of myoblast markers by TGF-beta2. J. Cell. Physiol. 196, 70–78 [DOI] [PubMed] [Google Scholar]
- 33.Schabort E. J., van der Merwe M., Loos B., Moore F. P., Niesler C. U. (2009) TGF-beta's delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp. Cell Res. 315, 373–384 [DOI] [PubMed] [Google Scholar]
- 34.Zentella A., Massagué J. (1992) Transforming growth factor beta induces myoblast differentiation in the presence of mitogens. Proc. Natl. Acad. Sci. U.S.A. 89, 5176–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Angelis L., Borghi S., Melchionna R., Berghella L., Baccarani-Contri M., Parise F., Ferrari S., Cossu G. (1998) Inhibition of myogenesis by transforming growth factor beta is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 95, 12358–12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn R. D., van Erp C., Habashi J. P., Soleimani A. A., Klein E. C., Lisi M. T., Gamradt M., ap Rhys C. M., Holm T. M., Loeys B. L., Ramirez F., Judge D. P., Ward C. W., Dietz H. C. (2007) Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat. Med. 13, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLennan I. S., Koishi K. (1997) Cellular localisation of transforming growth factor-beta 2 and -beta 3 (TGF-beta2, TGF-beta3) in damaged and regenerating skeletal muscles. Dev. Dyn. 208, 278–289 [DOI] [PubMed] [Google Scholar]
- 38.Budasz-Swiderska M., Jank M., Motyl T. (2005) Transforming growth factor-beta1 upregulates myostatin expression in mouse C2C12 myoblasts. J. Physiol. Pharmacol. 56, Suppl. 3, 195–214 [PubMed] [Google Scholar]
- 39.Smith C. A., Stauber F., Waters C., Alway S. E., Stauber W. T. (2007) Transforming growth factor-beta following skeletal muscle strain injury in rats. J. Appl. Physiol. 102, 755–761 [DOI] [PubMed] [Google Scholar]
- 40.Droguett R., Cabello-Verrugio C., Riquelme C., Brandan E. (2006) Extracellular proteoglycans modify TGF-beta bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biol. 25, 332–341 [DOI] [PubMed] [Google Scholar]
- 41.Brandan E., Cabello-Verrugio C., Vial C. (2008) Novel regulatory mechanisms for the proteoglycans decorin and biglycan during muscle formation and muscular dystrophy. Matrix Biol. 27, 700–708 [DOI] [PubMed] [Google Scholar]
- 42.Stewart C. E., Rotwein P. (1996) Insulin-like growth factor-II is an autocrine survival factor for differentiating myoblasts. J. Biol. Chem. 271, 11330–11338 [DOI] [PubMed] [Google Scholar]
- 43.Mourkioti F., Rosenthal N. (2005) IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 26, 535–542 [DOI] [PubMed] [Google Scholar]
- 44.Palsgaard J., Brown A. E., Jensen M., Borup R., Walker M., De Meyts P. (2009) Insulin-like growth factor I (IGF-I) is a more potent regulator of gene expression than insulin in primary human myoblasts and myotubes. Growth Horm. IGF Res. 19, 168–178 [DOI] [PubMed] [Google Scholar]
- 45.Rajpathak S. N., Gunter M. J., Wylie-Rosett J., Ho G. Y., Kaplan R. C., Muzumdar R., Rohan T. E., Strickler H. D. (2009) The role of insulin-like growth factor-I and its binding proteins in glucose homeostasis and type 2 diabetes. Diabetes Metab. Res. Rev. 25, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bach L. A., Headey S. J., Norton R. S. (2005) IGF-binding proteins–the pieces are falling into place. Trends Endocrinol. Metab. 16, 228–234 [DOI] [PubMed] [Google Scholar]
- 47.Duan C., Xu Q. (2005) Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 142, 44–52 [DOI] [PubMed] [Google Scholar]
- 48.Kumar A., Mohan S., Newton J., Rehage M., Tran K., Baylink D. J., Qin X. (2005) Pregnancy-associated plasma protein-A regulates myoblast proliferation and differentiation through an insulin-like growth factor-dependent mechanism. J. Biol. Chem. 280, 37782–37789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.James P. L., Jones S. B., Busby W. H., Jr., Clemmons D. R., Rotwein P. (1993) A highly conserved insulin-like growth factor-binding protein (IGFBP-5) is expressed during myoblast differentiation. J. Biol. Chem. 268, 22305–22312 [PubMed] [Google Scholar]
- 50.Salih D. A., Tripathi G., Holding C., Szestak T. A., Gonzalez M. I., Carter E. J., Cobb L. J., Eisemann J. E., Pell J. M. (2004) Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc. Natl. Acad. Sci. U.S.A. 101, 4314–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee A., Wilson E. M., Rotwein P. (2008) Insulin-like growth factor (IGF) binding protein-5 blocks skeletal muscle differentiation by inhibiting IGF actions. Mol. Endocrinol. 22, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren H., Yin P., Duan C. (2008) IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. J. Cell Biol. 182, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. (2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762–772 [DOI] [PubMed] [Google Scholar]
- 54.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 55.Florini J. R., Magri K. A., Ewton D. Z., James P. L., Grindstaff K., Rotwein P. S. (1991) “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 266, 15917–15923 [PubMed] [Google Scholar]
- 56.de Wit J., Verhaagen J. (2003) Role of semaphorins in the adult nervous system. Prog. Neurobiol. 71, 249–267 [DOI] [PubMed] [Google Scholar]
- 57.Roth L., Koncina E., Satkauskas S., Crémel G., Aunis D., Bagnard D. (2009) The many faces of semaphorins: from development to pathology. Cell Mol. Life Sci. 66, 649–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H., Wang X., Liu S., Wu Y., Zhao T., Chen X., Zhu L., Wu Y., Ding X., Peng X., Yuan J., Wang X., Fan W., Fan M. (2007) Sema4C participates in myogenic differentiation in vivo and in vitro through the p38 MAPK pathway. Eur. J. Cell Biol. 86, 331–344 [DOI] [PubMed] [Google Scholar]
- 59.Tatsumi R., Sankoda Y., Anderson J. E., Sato Y., Mizunoya W., Shimizu N., Suzuki T., Yamada M., Rhoads R. P., Jr., Ikeuchi Y., Allen R. E. (2009) Possible implication of satellite cells in regenerative motoneuritogenesis: HGF upregulates neural chemorepellent Sema3A during myogenic differentiation. Am. J. Physiol. Cell Physiol. 297, C238–C252 [DOI] [PubMed] [Google Scholar]
- 60.Delorme G., Saltel F., Bonnelye E., Jurdic P., Machuca-Gayet I. (2005) Expression and function of semaphorin 7A in bone cells. Biol. Cell 97, 589–597 [DOI] [PubMed] [Google Scholar]
- 61.McLoon L. K. (2009) A new role for satellite cells: control of reinnervation after muscle injury by semaphorin 3A. Focus on “Possible implication of satellite cells in regenerative motoneuritogenesis: HGF upregulates neural chemorepellent Sema3A during myogenic differentiation”. Am. J. Physiol. Cell Physiol. 297, C227–C230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y., Gunput R. A., Pasterkamp R. J. (2008) Semaphorin signaling: progress made and promises ahead. Trends Biochem. Sci. 33, 161–170 [DOI] [PubMed] [Google Scholar]
- 63.Svensson A., Libelius R., Tågerud S. (2008) Semaphorin 6C expression in innervated and denervated skeletal muscle. J. Mol. Histol. 39, 5–13 [DOI] [PubMed] [Google Scholar]
- 64.Adams R. H., Lohrum M., Klostermann A., Betz H., Püschel A. W. (1997) The chemorepulsive activity of secreted semaphorins is regulated by furin-dependent proteolytic processing. EMBO J. 16, 6077–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christensen C., Ambartsumian N., Gilestro G., Thomsen B., Comoglio P., Tamagnone L., Guldberg P., Lukanidin E. (2005) Proteolytic processing converts the repelling signal Sema3E into an inducer of invasive growth and lung metastasis. Cancer Res. 65, 6167–6177 [DOI] [PubMed] [Google Scholar]
- 66.Gonthier B., Nasarre C., Roth L., Perraut M., Thomasset N., Roussel G., Aunis D., Bagnard D. (2007) Functional interaction between matrix metalloproteinase-3 and semaphorin-3C during cortical axonal growth and guidance. Cereb. Cortex 17, 1712–1721 [DOI] [PubMed] [Google Scholar]
- 67.Gonthier B., Koncina E., Satkauskas S., Perraut M., Roussel G., Aunis D., Kapfhammer J. P., Bagnard D. (2009) A PKC-dependent recruitment of MMP-2 controls semaphorin-3A growth-promoting effect in cortical dendrites. PloS One 4, e5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bani C., Lagrota-Candido J., Pinheiro D. F., Leite P. E., Salimena M. C., Henriques-Pons A., Quirico-Santos T. (2008) Pattern of metalloprotease activity and myofiber regeneration in skeletal muscles of mdx mice. Muscle Nerve 37, 583–592 [DOI] [PubMed] [Google Scholar]
- 69.Nishimura T., Nakamura K., Kishioka Y., Kato-Mori Y., Wakamatsu J., Hattori A. (2008) Inhibition of matrix metalloproteinases suppresses the migration of skeletal muscle cells. J. Muscle Res. Cell Motil. 29, 37–44 [DOI] [PubMed] [Google Scholar]
- 70.Elhabazi A., Delaire S., Bensussan A., Boumsell L., Bismuth G. (2001) Biological activity of soluble CD100. I. The extracellular region of CD100 is released from the surface of T lymphocytes by regulated proteolysis. J. Immunol. 166, 4341–4347 [DOI] [PubMed] [Google Scholar]
- 71.Basile J. R., Holmbeck K., Bugge T. H., Gutkind J. S. (2007) MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. J. Biol. Chem. 282, 6899–6905 [DOI] [PubMed] [Google Scholar]
- 72.Dean R. A., Overall C. M. (2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 73.Kaelin W. G. (2005) Proline hydroxylation and gene expression. Annu. Rev. Biochem. 74, 115–128 [DOI] [PubMed] [Google Scholar]
- 74.Krane S. M. (2008) The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids 35, 703–710 [DOI] [PubMed] [Google Scholar]
- 75.Kratchmarova I., Kalume D. E., Blagoev B., Scherer P. E., Podtelejnikov A. V., Molina H., Bickel P. E., Andersen J. S., Fernandez M. M., Bunkenborg J., Roepstorff P., Kristiansen K., Lodish H. F., Mann M., Pandey A. (2002) A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol. Cell. Proteomics 1, 213–222 [DOI] [PubMed] [Google Scholar]
- 76.Saltiel A. R. (2001) You are what you secrete. Nat. Med. 7, 887–888 [DOI] [PubMed] [Google Scholar]
- 77.Pedersen B. K. (2009) Edward F. Adolph distinguished lecture: muscle as an endocrine organ: IL-6 and other myokines. J. Appl. Physiol. 107, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 78.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57, 125–133 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.