Abstract
Although the genome of the Mycobacterium tuberculosis H37Rv laboratory strain has been available for over 10 years, it is only recently that genomic information from clinical isolates has been used to generate the hypothesis of virulence differences between different strains. In addition, the relationship between strains displaying differing virulence in an epidemiological setting and their behavior in animal models has received little attention. The potential causes for variation in virulence between strains, as determined by differential protein expression, have similarly been a neglected area of investigation. In this study, we used a label-free quantitative proteomics approach to estimate differences in protein abundance between two closely related Beijing genotypes that have been shown to be hyper- and hypovirulent on the basis of both epidemiological and mouse model studies. We were able to identify a total of 1668 proteins from both samples, and protein abundance calculations revealed that 48 proteins were over-represented in the hypovirulent isolate, whereas 53 were over-represented in the hypervirulent. Functional classification of these results shows that molecules of cell wall organization and DNA transcription regulatory proteins may have a critical influence in defining the level of virulence. The reduction in the presence of ESAT-6, other Esx-like proteins, and FbpD (MPT51) in the hypervirulent strain indicates that changes in the repertoire of highly immunogenic proteins can be a defensive process undertaken by the virulent cell. In addition, most of the previously well characterized gene targets related to virulence were found to be similarly expressed in our model. Our data support the use of proteomics as a complementary tool for genomic comparisons to understand the biology of M. tuberculosis virulence.
Comparative genomics has shown that the global epidemic of TB1 is a composite of a myriad of different strains that can be grouped into phylogenetic lineages according to the presence or absence of specific genetic markers (1). The frequency at which strains representative of the different lineages occur within a community and/or globally is thought to be a marker of their level of fitness. In part, this is reflected in animal model infection studies that show that different strains have different levels of virulence, measured by the rate at which they are able to kill the animal host (2).
Immunological investigation noted that different strains elicit different helper responses during the early phase of infection (3). The CDC1551 Mycobacterium tuberculosis strain was shown to induce a protective Th1 response, whereas certain Beijing genotype strains were found to induce a non-protective Th2 response (4). The mechanism(s) underlying the pathogenic characteristics of different strains remains largely unknown with the exception of the region of difference RD1, which encodes for the virulence proteins ESAT-6 and CFP-10 in all strains (5). Other possible virulence factors include the two-component regulatory (2CR) molecules with the dosR gene as one example (6) as well as less characterized deletions such as the stf3 (7) and the putative transcriptional regulator Rv1773 (8) among others.
Within the Beijing lineage, it has been demonstrated that the phenolic glycolipid (PGL), a product of the pks15 gene product, inhibits secretion of TNF-α, interleukin-6, and interleukin-12, thereby modulating the host immune system (9). However, complementation of H37Rv with the pks 15 gene was unable to induce a higher level of virulence, demonstrating that additional virulence factors must be involved (10). Unfortunately, whole genome sequencing has not identified a set of genes that can explain virulence in M. tuberculosis (11). It is evident from these studies that the M. tuberculosis genome continues to evolve by single nucleotide polymorphisms, insertions, and deletions as well as transposition of transposable elements (12–14). It is not known how these genetic changes translate to virulence differences; however, identifying protein expression and protein abundance differences through transcriptomics and proteomics approaches may increase our understanding of M. tuberculosis biology and virulence.
The ability to characterize complex proteomes in depth has been dramatically increased by technological developments in mass spectrometry-based proteomics (15). In particular, a hybrid mass spectrometer, the linear ion trap-Orbitrap (16, 17), allows faster mass measurements, whereas acquisition methods ensure enhanced coverage of the sample. In addition, high mass accuracy can routinely be achieved in both the MS and MS/MS mode by using a “lock mass” strategy (18) where ions present in the air and thereby in every measurement can be used as an internal standard, which virtually eliminates the problem of false-positive peptide identifications. However, mass spectrometry-based approaches were initially incapable of providing quantitative information, which could only be obtained with the incorporation of stable isotopic labeling in the samples through metabolic or chemical methods (19, 20). Unfortunately, most chemical methods use isotopic tags that can only be efficiently analyzed by certain MS/MS instrumentations, and metabolic labeling can be very challenging for an organism with very low metabolic rates in culture as is the case of M. tuberculosis. Therefore, the use of protein abundance calculations that do not require peptide labeling is desirable. One such label-free approach, exponentially modified protein abundance index (emPAI) (21) can be done by using the observable number of parent ions from a certain protein and the theoretical number of peptides expected for the same protein. Such estimates have been shown to correlate relatively precisely with individual protein concentrations within a sample (21, 22).
In this study, we used Orbitrap technology in combination with emPAI calculations to describe proteomic differences between two closely related Beijing genotype strains. These strains have vastly different pathogenic characteristics in terms of their ability to transmit and cause disease in humans and to cause pulmonary damage in mice.
EXPERIMENTAL PROCEDURES
Genotypic Classification of M. tuberculosis Isolates
M. tuberculosis isolates were cultured from TB patients attending primary health care clinics in South Africa. Cultures positive for M. tuberculosis were genotyped by IS6110 DNA fingerprinting and spoligotyping using the internationally standardized methods. Transmission chains were defined as a series of cases having isolates with identical IS6110 DNA fingerprints with intercase intervals of less than 2 years, and each transmission chain was assumed to be initiated by a single index case (23). A transmission chain-unique case was defined as one having no other cases with the identical strain occurring within 2 years either side (23). Strains with the Beijing genotype were identified by their characteristic spoligotype and were grouped into phylogenetic sublineages as described previously (24).
Experimental Model of Pulmonary Tuberculosis in BALB/c Mice
Virulence (defined by survival, lung pathology, and bacterial load) induced by each selected isolate was evaluated in 6–8-week-old male BALB/c mice as described previously (2, 25–29). Briefly, bacteria were grown in Middlebrook 7H9 broth (Difco) enriched with glycerol, albumin, catalase, and dextrose (BD Biosciences) and incubated with constant agitation at 37 °C and 5% CO2 for 21 days. Growth was monitored by densitometry. As soon as the culture reached midlog growth phase (A600 = 1) the bacilli were harvested, the concentration was adjusted to 2.5 × 105 viable bacilli/100 μl of PBS as determined by diacetate fluorescein incorporation, and 100 μl aliquots were frozen at −70 °C until use. To induce progressive pulmonary tuberculosis, mice were anesthetized with sevoflurane and inoculated intratracheally with 2.5 × 105 bacilli in 100 μl of PBS (26, 27). Infected mice were kept in a vertical position until the effect of anesthesia passed.
Two experiments were performed: in each experiment two groups of 70 mice were infected with the two different clinical M. tuberculosis strains. Twenty mice from each group were left undisturbed to record survival up to day 120 after infection. Six animals from each group were sacrificed by exsanguination at each of 1, 3, 7, 14, 21, 28, 60, and 120 days after infection. One lung lobe, right or left, was perfused with 10% formaldehyde (dissolved in PBS), and samples were prepared for histopathology, determining by automated morphometry the percentage of lung surface area affected by pneumonia. The other lobe was snap frozen in liquid nitrogen and used for the determination of bacillary load by counting the number of colony-forming units (cfu) following the method described previously (26, 27). All procedures were performed in a class III cabinet in a biosafety level III facility. Infected mice were kept in cages fitted with microisolators connected to negative pressure. Animal work was performed in accordance with the national regulations on animal care and experimentation in Mexico.
Mycobacterial Culturing
Stock cultures of two genetically closely related Beijing genotype strains that demonstrate vastly different pathogenic characteristics in terms of their ability to transmit and cause disease in human and to kill mice were inoculated into mycobacterial growth indicator tubes and incubated at 37 °C until positive growth was detected using the Bactec 460 TB system (BD Biosciences). Approximately 0.2 ml was inoculated onto Löwenstein-Jensen medium and incubated at 37 °C over 6 weeks with weekly aeration until colony formation. Colonies were transferred into 20 ml of supplemented 7H9 Middlebrook medium (BD Biosciences) containing 0.2% (v/v) glycerol (Merck Laboratories), 0.1% Tween 80 (Merck Laboratories), and 10% dextrose, catalase and incubated at 37 °C. Once the culture reached an A600 of 0.9, 1 ml was inoculated into 80 ml of supplemented 7H9 Middlebrook medium and incubated at 37 °C until an A600 between 0.6 and 0.7 was reached. M. tuberculosis cells in this midlog growth phase were used for whole cell lysate protein extractions.
Preparation of Crude Mycobacterial Extracts
Mycobacterial cells were collected by centrifugation (10 min at 2500 × g) at 4 °C and resuspended in 1 ml of cold lysis buffer containing 10 mm Tris-HCl, pH 7.4 (Merck Laboratories), 0.1% Tween 80 (Sigma-Aldrich), one tablet/25 ml Complete protease inhibitor mixture (Roche Applied Science), and one tablet/10 ml phosphatase inhibitor mixture (Roche Applied Science). Cells were transferred into 2 ml cryogenic tubes with O-rings, and the pellet was collected after centrifugation (5 min at 6000 × g) at 21 °C. An equal volume of 0.1 mm glass beads (Biospec Products Inc., Bartlesville, OK) was added to the pelleted cells. In addition, 300 μl of cold lysis buffer including 10 μl of 2 units/ml RNase-free DNase I (New England Biolabs) was added, and the cell walls were lysed mechanically by bead beating for 20 s in a Ribolyser (Bio101 Savant, Vista, CA) at a speed of 6.4. Thereafter the cells were cooled on ice for 1 min. The lysis procedure was repeated three times. The lysate was clarified by centrifugation (10,000 × g for 5 min) at 21 °C, and the supernatant containing the whole cell lysate proteins was retained. Thereafter the lysate was filter-sterilized through a 0.22 μm-pore Acrodisc 25 mm PF syringe sterile filter (Pall Life Sciences, Pall Corp., Ann Arbor, MI) and stored at −80 °C until further analysis.
Gel Electrophoresis and In-gel Trypsin Digestion
Whole cell lysate protein was diluted in HPLC grade water to obtain a final concentration of 4 μg/μl. Fifty-two micrograms of the protein (13 μl) was added to an application buffer mixture containing 2 μl of HPLC grade water, 5 μl of sample buffer, and 1 μl of DTT and heated for 5 min at 65 °C. Thereafter the proteins were fractionated in duplicate by SDS-PAGE using a 4–12% gradient, 1.0 mm NuPAGE gel (Invitrogen) under reducing conditions for 40 min at 175 V. SDS-PAGE gels were Coomassie stained using a Colloidal Blue Staining kit for NuPAGE. After staining, each gel lane was divided into 12 fractions, and each fraction was subjected to in-gel reduction, alkylation, and tryptic digestion. In brief, proteins were reduced using 10 mm DTT for 1 h at 56 °C and alkylated with 55 mm iodoacetamide for 45 min at room temperature. The reduced and alkylated peptides were digested with sequence grade, modified trypsin 1:50 (w/w) (Promega, Madison, WI) for 16 h at 37 °C in 50 mm NH4HCO3, pH 8.0. The reaction was quenched through acidification with 2% TFA (Fluka, Buchs, Switzerland). The resulting peptide mixture was desalted on reverse phase C18 stop and go extraction tips (30) and diluted in 0.1% TFA prior to nano-HPLC-MS analysis.
Mass Spectrometry
All experiments were performed on a Dionex Ultimate 3000 nano-LC system (Dionex, Sunnyvale, CA) connected to a linear quadrupole ion trap-Orbitrap (LTQ-Orbitrap) mass spectrometer (Thermo Electron, Bremen, Germany) equipped with a nanoelectrospray ion source. For liquid chromatography separation, we used an Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex) capillary of 12 cm bed length and 100 μm inner diameter self-packed with ReproSil Pur C18-AQ (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The flow rate used was 0.3 μl/min for the nanocolumn, and the solvent gradient used was 7–40% B in 87 min and then 40–80% B in 8 min. Solvent A was aqueous 2% ACN in 0.1% formic acid, and solvent B was aqueous 90% ACN in 0.1% formic acid.
The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full-scan MS spectra (from m/z 300 to 2000) were acquired in the Orbitrap with resolution of R = 60,000 at m/z 400 (after accumulation to a target of 1,000,000 charges in the LTQ). The method used allowed sequential isolation of the most intense ions (up to six, depending on signal intensity) for fragmentation on the linear ion trap using collisionally induced dissociation at a target value of 100,000 charges.
For accurate mass measurements, the lock mass option was enabled in MS mode, and the polydimethylcyclosiloxane ions generated in the electrospray process from ambient air were used for internal recalibration during the analysis (18). Target ions already selected for MS/MS were dynamically excluded for 60 s. General mass spectrometry conditions were as follows: electrospray voltage, 1.5 kV; no sheath; and auxiliary gas flow. Ion selection threshold was 500 counts for MS/MS, and an activation Q-value of 0.25 and activation time of 30 ms were also applied for MS/MS.
Mascot Search and Peptide/Protein Validation
MS/MS peak lists from individual 48 RAW files, 24 from the hypovirulent and 24 from the hypervirulent strains, were generated using the DTA SuperCharger package, version 1.29, using default extract_msn parameters, available in the MSQuant validation tool (see below). Protein identification was performed by searching the data separately against M. tuberculosis H37Rv protein database version R11 available at the Tuberculist website (genolist.pasteur.fr/tuberculist/) containing 4066 entries. The databases were in-house modified to also contain reversed sequences of all entries as a control of false-positive identifications during analysis (total number of entries in the database, 8162). Common contaminants, such as keratins, BSA, and trypsin, were also added to the database. We used Mascot Daemon for multiple search submission on a local Mascot server v2.1 (Matrix Science). The search parameters used were as follows: enzyme specificity, trypsin/with no proline restriction; maximum missed cleavages, 3; carbamidomethyl (Cys) as fixed modification; N-acetyl (protein), oxidation (Met), pyro-Glu (Gln), and pyro-Glu (Glu) as variable modifications; precursor ion mass tolerance, 15 ppm; and MS/MS mass tolerance, 0.5 Da. Under these criteria, Mascot indicated a minimal score of 22 for p ≤ 0.01 and 15 for p ≤ 0.05. All data had a mass accuracy average of 4.1 ppm. Spectrum and protein validation was performed using an open source software called MSQuant (version 1.5a61) largely used for LC-MS/MS data analysis (31). Proteins were validated statistically based on the score of their individual peptides. Proteins with at least two tryptic peptides with a minimal score of 22 for each (protein false-positive probability of 0.01%) or those with only one peptide but a MS/MS score higher than 38 were accepted (protein false-positive probability lower than 0.25%). Using these criteria, all MS/MS identifications of peptides present in entries with reversed sequences (i.e. false-positive identifications) were not validated because none of the reversed proteins were identified with two peptides with a score higher than 21 each or one peptide with a score higher than 38 (the highest Mascot score for a peptide from the reversed database was 32; data not shown). Identifications with only one unique peptide were accepted only after manual validation. Redundant peptides shared by a protein family were reported only once for the family member with the best scoring in the search result. Subsequent members of the protein family were only reported if a unique peptide was identified. If no additional unique peptides were present, only one member of the family was considered. Quality criteria for manual validation were the assignment of major peaks, the occurrence of uninterrupted y or b ion series of at least three consecutive amino acids, the preferred cleavages amino-terminal to proline bonds and carboxyl-terminal to Asp or Glu bonds, and the possible presence of a2/b2 ion pairs. All MS/MS fragmentation patterns (peak lists) are publicly available at the Tranche Network (proteomecommons.org) through the hash code iTQNvA0gKr5jo8Smv6o96qLlpbCINzlpVb7lbz7LKh24SuyLUzTnorzk/5wuI2Qyuh4sOP5Eqjd7xmr1oHg8XivhLKoAAAAAAAAB6g==.
Protein Abundance Estimation
Protein abundance expressed as emPAI values was calculated using the number of observable peptides and the number of observed parent ions per identified peptide. The number of observable (or expected) peptides for a protein was calculated through in silico trypsin digestion of the M. tuberculosis H37Rv database, and the resulting peptide fragments were compared with the scan range of the mass spectrometry. The emPAI values were calculated using a script developed at the Keio University (empai.iab.keio.ac.jp) using the following parameters: trypsin as enzyme, carbamidomethyl (Cys) fixed modification, mass range from 300 to 8000 Da, no retention time filtering, bold red peptides only (i.e. unique peptides in the Mascot result), and peptides filtered by peptide Mascot score higher than 21. The protein abundance index (PAI) was obtained by division of the observed parent ions by the number of theoretical observable peptides; emPAI was obtained using the formula emPAI = 10PAI − 1. To obtain the concentration of a protein in the sample, its emPAI value was divided by the sum of all emPAI values in the sample, and the result was multiplied by 100, thus resulting in an estimate of the mol % of the protein (21).
To determine differentially represented proteins between the hypo- and the hypervirulent M. tuberculosis strains, we merged the peptide list identifications of two independent replicates per strain. Individual mol % values were compared, and proteins were divided in two categories as follows. (i) For proteins identified in both samples, the difference in relative concentration between the strains had to be higher than 4-fold. (ii) For a protein identified in only one of the strains, we required that it had to be identified with a minimum of three parent ions, and we selected a mol % of 0.02 as a threshold. Such stringent criteria are required to guarantee that a protein identified in only one sample is most probably due to differences in abundance between the samples and not because parent ions were not identified (but still present) in the MS analysis due to random fluctuation of the MS/MS data-dependent acquisition procedure.
Quantification of CFP-10 and ESAT-6 Gene Expression by Real Time PCR during Murine Pulmonary Tuberculosis
We used the previously described technique with some modification to determine mycobacterial gene expression in infected mice lungs (32, 33). In brief, lungs from three mice for each time point were perfused with 1 ml of TRIzol (Qiagen, Valencia, CA). The tissue was minced, transferred to a sterile tube, and homogenized using a Multi-Gen 7 mm homogenizer for three cycles at maximal speed with 30 s bursts. The homogenized sample was centrifuged at 4000 rpm for 10 min at 20 °C, and the supernatant was transferred to a new tube and immediately place on ice. This supernatant contains eukaryotic RNA, whereas the pelleted material contains the bacilli. To isolate the bacterial RNA, 1 ml of TRIzol was added to this pellet, and the pellet was resuspended and transferred to a 2 ml screw cap tube containing zirconia and silica beads (diameter, 1 and 0.1 mm, respectively). Mycobacteria were disrupted using a Mini-Bead-Beater-8 apparatus (Biospec Products Inc.) using three cycles of 30 s each at the highest speed. This material was then centrifuged for 1 min at maximum speed, and the supernatant was transferred to a 2 ml screw cap tube containing 300 μl of chloroform. After vigorous vortexing and centrifugation for 10 min at maximum speed, the resulting supernatant contained M. tuberculosis RNA, which was precipitated with a solution of glycogen, ammonium acetate, isopropanol, and ethanol. Bacterial RNA was isolated using four cycles of purification with an RNeasy column (Invitrogen) followed by extensive DNase incubation to eliminate DNA contamination.
We used the mycobacterial 16 S ribosomal RNA for reference constitutive gene expression. Primers for 16 S, cfp-10, and esat-6 genes were designed with the Primer Express software, version 2.0 (Applied Biosystems). The nucleotide sequences of the forward and the reverse primers were as follows: 16 S forward, 5′-tcccgggccttgtacaca-3′; 16 S reverse, 5′-ccactggcttcgggtgtta-3′; cfp-10 forward, 5′-gatgaagaccgatgccgcta-3′; cfp-10 reverse, 5′-cgctcgaaattacctgcctc-3′; esat-6 forward, 5′-accagggtgtccagcaaaaa-3′; and esat-6 reverse, 5′-cgttgttcagctcggtagcc-3′.
The quality and quantity of RNA were evaluated by spectrophotometric measurements (260/280 nm) and on agarose gels. cDNA synthesis was performed using 5 μg of total RNA, 2 μm random primers (Promega), 10 units/μl ribonuclease inhibitor (Invitrogen), 1× Buffer RT, 0.5 mm each dNTP, and 4 units of Omniscript reverse transcriptase (Qiagen). A preliminary conventional PCR using 16 S ribosomal RNA primers was carried out with an aliquot of cDNA. The PCR product was used for quantification on an agarose gel. Concentration was determined by comparing the intensities of fluorescence of the 16 S fragment with the SYBR Green-stained DNA mass ladder fragments of known concentration (Invitrogen). Real time quantitative PCR was performed with the QuantiTect SYBR Green PCR Master Mix (Qiagen). To obtain a standard curve, four different PCRs were performed in parallel using as template 10-fold dilutions of known amounts of the M. tuberculosis H37Rv 16 S rRNA gene (108–102 copies) together with the experimental sample. Reactions were performed in sealed tubes in a 96-well microtiter plate in a 7500 Prism spectrofluorometric thermal cycler (Applied Biosystems) in a volume of 20 μl. The reaction mixtures consisted of 0.5 μm target and control primers, 12.5 μl of Master Mix, and 1 μg of cDNA. Cycling conditions were as follows: initial denaturation for 15 min at 95 °C followed by 40 cycles at 95 °C for 20 s, 59 °C for 20 s, and 72 °C for 34 s. An independent real time PCR assay was carried out in triplicate for each mouse lung (at least three mice per time point) in two separate experiments. The mRNA copy number of mycobacterial genes was related to one million copies of RNA encoding the 16 S rRNA gene. PCR fragments amplified from Rv1759c and 16 S rRNA gene from M. tuberculosis were identified by means of a genetic analyzer, ABI PRISM 310 PE (Applied Biosystems).
RESULTS
Molecular Epidemiology
Genotyping of M. tuberculosis from our strain collection identified 452 isolates with the Beijing genotype of which 319 were classified as members of sublineage 7 according to 40 different genetic markers (24). Within this sublineage, 288 cases were clustered into 21 transmission chains (cluster size ranged from 2 to 147), and 31 cases had unique IS6110 DNA fingerprints. To determine whether an actively transmitting strain showed pathogenic properties different from that of a non-transmitting strain, one isolate representative of the largest cluster (n = 147 cases) and one unique isolate were randomly selected for further analysis (Fig. 1).
Fig. 1.
IS6110 DNA fingerprint of selected Beijing genotype strains. A, strain representative of the largest cluster (hypervirulent). B, strain that did not transmit during the study period (hypovirulent).
Survival, Lung Histopathology, and Bacillary Loads in BALB/c Mice Infected by Intratracheal Route with Selected Beijing Strains
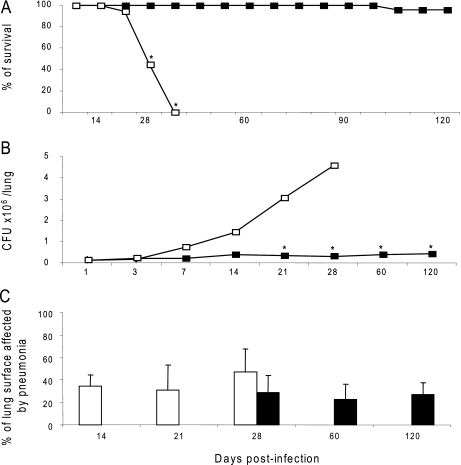
Mice infected with the actively transmitting strain started to die 3 weeks after infection, and by 5 weeks, all the mice had died (Fig. 2A). Culture of lung homogenates showed very high lung bacillary loads (Fig. 2B), and histopathology showed extensive tissue damage (pneumonia) that started at day 14 postinfection (Fig. 2C). Based on these results we assigned a virulence status of hypervirulent to this Beijing genotype. Animals infected with the unique strain showed 90% survival after 4 months of infection (Fig. 2A), 10-fold lower cfu in the lungs (Fig. 2B), and reduced tissue damage (pneumonia) that started at day 28 postinfection (Fig. 2C). Based on these results we assigned a virulence status of hypovirulent to this Beijing strain. Because the growth of the hypovirulent strain seems to be atypical for a M. tuberculosis strain, we also performed an in vitro growth curve of both strains. Supplemental Fig. 1 shows that, up to 30 days after the culture was established, both strains grew at similar rates.
Fig. 2.
Virulence of M. tuberculosis clinical isolates in murine infection model. BALB/c mice were infected by intratracheal injection (2.5 × 105 bacilli) with selected Beijing strains. The virulence of the strain representative of the largest cluster (white symbols) was compared with the virulence of the non-transmitting strain (black symbols). A, Kaplan-Meier survival curves. B, bacillary loads. C, morphometry (percentage of lung surface area affected by pneumonia). Asterisks represent statistical significance (p < 0.005). Error bars indicate 95% confidence intervals.
Proteome Data
To identify differences in abundance of proteins in the two Beijing genotype strains, whole cell lysate proteins were subjected to the gel-LC-MS/MS approach (15). The electrophoretic protein profile of both strains is almost identical (data not shown). All mass spectrometry data obtained were identified using Mascot and were submitted for identification against the M. tuberculosis H37Rv laboratory strain protein database (Tuberculist). This reference database was used because previous studies have suggested that this gene annotation is the most reliable (34).
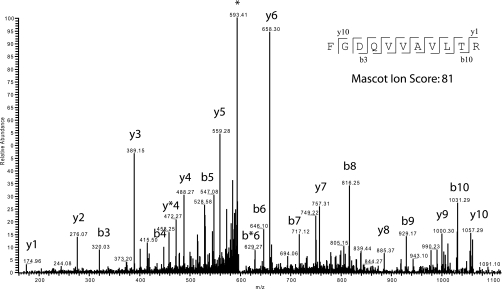
Fig. 3 illustrates an example of an MS/MS spectrum of a peptide of m/z 602.84. The Mascot tool identified it as the peptide FGDQVVAVLTR, corresponding to the protein entry Rv3220c, annotated as “probable two-component sensor kinase.” This identification had a mass accuracy of 4.4 ppm between observed and theoretical masses. Fig. 3 also illustrates the fragmentation pattern and identification of y/b ion series of the sequence (sequence input). In this example, Mascot was able to correlate the full y ion series and eight of 10 b ions, resulting in a Mascot identification score of 81 (a score of 22 represents a p < 0.01).
Fig. 3.
MS/MS profile of ion m/z 601.41. A tandem mass spectrum of a prevalent ion on a particular time point in the LC gradient and ionized on the LTQ-Orbitrap is shown. The peptide fragments randomly on each amide bond, resulting in carboxyl-terminal y ions or amino-terminal b ions. After the fragment masses were submitted to Mascot, the peptide was identified as FGDQVVAVLTR (inset with detected y and b ions represented) from protein Rv3220c, a probable two-component sensor kinase. * Asterisk represent the parent ion.
In summary, the full analysis of both replicates for each sample identified a total of 1440 proteins for the hypovirulent strain and 1521 proteins for the hypervirulent strain. Merging both data sets, we identified the protein products of 1668 of the 4066 genes predicted by Tuberculist. From these, 145 proteins were identified only in the hypovirulent cells, and 226 were identified only in the hypervirulent strain. The majority of these proteins observed in only one strain represent identifications based on one or two peptides, and in these cases, those proteins were not used for quantitative comparison. Supplemental File S1 reports all peptides identified in whole lysate protein extract from the two Beijing genotype strains, their protein mass, number of peptides per identified protein, peptide length, observed charge, observed m/z ratio, measured peptide mass (Da), Mascot score, the presence of modifications (such as amino-terminal acetylation or Met oxidation), and the error of the observed/theoretical mass in ppm.
emPAI Analysis for Quantitative Protein Estimation and Data Comparison
The peptide list input (as shown in supplemental File S1) was submitted to the emPAI calculation tool, and emPAI values for individual proteins identified in each sample were obtained. Before the submission of the data, we manually merged proteins Rv1198, Rv1793, and Rv2346c and renamed the complex as ESATx-like. This was done because it is impossible to determine which of those proteins are contributing the tryptic peptides that are shared between them (these 94-amino acid-long proteins share 86 identical amino acid residues).
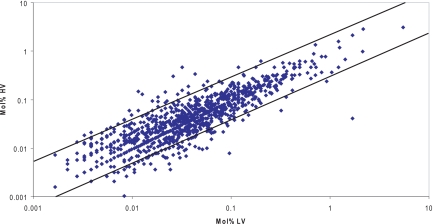
The protein concentration in the sample was calculated as mol % by dividing individual emPAI values by the sum of all values within a sample and multiplying by 100. This step is not only important for measuring the mol % of a protein but also to normalize any difference in emPAI values observed between samples due to differences in instrument efficiency during different runs. After normalization and mol % calculation, all mol % values from proteins present in both samples were plotted in a logarithmic linear distribution graph (Fig. 4). This analysis showed that most of the 1298 proteins lie within a difference range of less than 3-fold. This indicates not only that the normalization is reliable but also that the relative quantitation provided by emPAI is relatively accurate because the majority of proteins in this comparison are equally distributed between the two data sets. Supplemental File S2 contains all emPAI values obtained for each strain and a sheet with the merged results for both strains.
Fig. 4.
Protein abundance comparison of hypo- and hypervirulent cells. Protein abundance was derived from emPAI values. Solid lines delimit differentially abundant proteins (external area) from similarly abundant (internal area). HV, high virulent; LV, low virulent.
Identification of Differentially Abundant Proteins
Once mol % was calculated and samples were compared, we set stringent thresholds to indicate proteins differentially represented in each Beijing strain as shown under “Experimental Procedures.” When these chosen criteria were applied to our data set, only 53 entries from the hypervirulent strain and 48 from the hypovirulent strain were selected. emPAI values for all entries can be seen in supplemental File S2, and a short description of these entries is also reported in supplemental File S3.
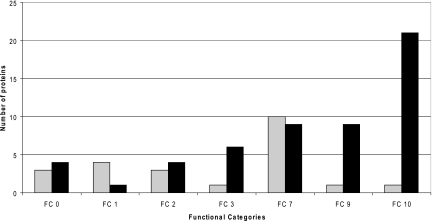
Fig. 5 shows these differential proteins grouped according to the functional category as given in Tuberculist. Interestingly, proteins in functional categories 3 (cell wall and cell processes) and 9 (regulatory proteins) are significantly over-represented in the hypervirulent strain, whereas proteins from functional category 1 (lipid metabolism) are more evident in the hypovirulent strain. However, because we did not perform any enrichment of membrane proteins, there is some uncertainty with the observed quantitative ratios. The proteins identified in each functional group and short descriptions of their functions are reported in supplemental File S3.
Fig. 5.
Functional classification of differentially abundant proteins. Proteins were classified according to the Tuberculist functional category (FC) groups. Gray bars represent proteins that are more abundant in the hypovirulent strain, and black bars represent proteins that are more abundant in the hypervirulent strain. FC10 represents the group of conserved hypothetical proteins.
In Vivo RT-PCR Measurements
Because ESAT-6 is an exported protein, intracellular accumulation of this protein demonstrated by proteomics could be a result of gene expression changes or changes in membrane transport machinery. Further validation of gene expression levels of ESAT-6 and CFP-10 was performed in vivo during several time points of the infection (supplemental Fig. 2). In supplemental Fig. 2A, quantitative information of ESAT-6 from mice infected with one or the other strain was normalized against the number of copies of 16 S rRNA gene used as a control. In supplemental Fig. 2B, the absolute value of gene copies is divided by 1 cfu. The results showed that ESAT-6 gene expression was reduced in the hypervirulent strain (supplemental Fig. 2B, white bars). CFP-10 showed a similar reduction but to a lesser extent (data not shown).
DISCUSSION
The identification of virulence factors from M. tuberculosis involved in the TB disease process is not only a crucial step to understanding the biology of the pathogen but may also provide insights into regional TB epidemiology that in turn may improve treatment regimens in the long run. The release of the M. tuberculosis H37Rv laboratory strain genome (11) and further availability of other genomes of the M. tuberculosis complex such as Mycobacterium bovis, the attenuated M. bovis BCG vaccine strain, and the clinical isolate M. tuberculosis CDC1551 have allowed many genomic comparisons to discover regions in the genome with gene mutations or gene deletions that result in a more or less virulent phenotype (35, 36). However, it can be expected that many factors participating in virulence of a strain cannot be directly identified at the genomic level, and information regarding gene expression and protein abundance can contribute greatly to improve our understanding in this field.
To test this hypothesis, we used an MS-based proteomics approach coupled with a label-free abundance estimate to identify proteins present in two clinical Beijing strains isolated from TB cases in South Africa. These isolates are of particular interest because they display striking differences in their level of virulence as defined by their epidemiological and population characteristics as well as virulence in a mouse infection model. The fact that the strain we defined as hypervirulent using epidemiological studies was also found to be hypervirulent using the definition of a rapid onset of disease and death in a mouse model is of itself an interesting and in some ways counterintuitive result. A common and long held view of pathogen evolution is that strains that rapidly kill their host may be negatively selected because the opportunities for transmission to new hosts are reduced. However, it is recognized that natural selection can still favor these strains as long as they possess a compensatory advantage, for example higher instantaneous transmissibility rates or increased resistance to host defenses (37). Thus, it is possible that this strain is able to overcome the selective disadvantage of rapid killing of the host via mechanism(s) such as these. We also recognize that because of the significant differences in TB pathology between inbred laboratory mice and humans (38) our animal model results are not necessarily translatable to our epidemiological results.
From a total of 1668 identified proteins, we applied very stringent criteria to select a minimal observed difference threshold, and in total, 101 proteins were identified as being significantly differentially abundant in either sample. Our criteria are in accordance with the reported accuracy of emPAI, which has been shown to have an error range close to a maximal factor of 3 (21). Those 101 proteins were clustered into three functional categories as defined in the Tuberculist resource database established by Cole et al. (11). In addition, the remaining proteins not classified as differentially abundant were also used to check the presence of proteins previously described as virulence markers or factors.
For example, one of the well characterized genomic regions associated with virulence is the phthiocerol dimycocerosate (PDIM) locus. Mutants containing deletions of this locus were unable to multiply in mouse models (39). Some members of the PDIM family are sometimes linked to a PGL molecule, and such glycolipids are found to be very characteristic of highly virulent Beijing strains (9). The genes that participate in PGL synthesis and modification are located together within the PDIM locus. In recent years, this locus has been analyzed in detail, and the functions of many of these genes have been determined (40). In this work, we identified peptides representing proteins from 18 genes of a total of 35 genes present in that locus. From those, only two proteins (PpsA and FadD28) were not observed in a 1:1 ratio, but still the calculated differences were below our stringent threshold, being 2.5- and 2.8-fold over-represented in the hypovirulent strain, respectively, showing that in principle PDIM and PGL synthesis are expected to be similar in this model.
Molecules involved in the regulation of transcription (functional category 9) could be the main factors regulating virulence in our model. This could be particularly relevant because if a single transcriptional regulator effects the expression of numerous genes (either directly or indirectly) a large phenotypic effect could result from a minor genetic change. Interestingly, the relevance of these genes in pathogenesis is illustrated by examples where the inactivation of genes encoding σ factors (41, 42) or 2CR systems (43, 44) causes attenuation of virulence in vivo. On the other hand, mutations or deletions in some of these 2CR systems also involve increased virulence (6). Strikingly, this was shown for the regulon devR in mouse models, whereas alteration in the same gene has opposite effects in other animal infection models (44). In addition, most of the gene targets for these transcriptional regulators remain unknown up to the present date. This further increases the challenge in assessing the participation of regulons in virulence acquisition. This type of knowledge is essential because some of these regulators might be involved in inducing or repressing immunostimulatory molecules (36). We identified 10 regulatory proteins, nine of which were over-represented in the hypervirulent strain. Of these, only Rv3574 has been investigated in depth. It represses the expression of an estimated 74 genes in M. tuberculosis (45), some of which are involved in lipid metabolism. It was shown that deletion of Rv3574 leads to attenuation of virulence, which is in accordance with our data. However, how this and other regulators actually participate in virulence itself is far from clear.
Well described immunogenic proteins such as ESAT-6, Esx-like proteins, and MPT51 (FbpD) (46) were under-represented in the hypervirulent strain. Interestingly, ESAT-6 and Esx-like proteins are highly abundant and considered well defined virulence factors in M. tuberculosis because the deletion of RD1 containing this gene results in loss of virulence (47) as observed for the attenuated M. bovis BCG strain (5). Previously, our group determined that the Esx-like proteins had a minimum 1.0 mol % in the avirulent M. tuberculosis H37Ra strain and up to 7.0 mol % in M. tuberculosis H37Rv (data not shown). However, the hypervirulent Beijing strain showed only 0.23 mol %. Nonetheless, it is important to note that although in this study we analyzed whole cell extracts the decreased levels of ESAT-6 and Esx-like proteins in the hypervirulent strain may be a result of faster export of these proteins to the extracellular environment, which is their normal localization. Therefore, we performed RT-PCR of ESAT-6 in vivo during the course of infection to better test that hypothesis. Our data show that ESAT-6 gene expression was indeed reduced in the hypervirulent cell. This protein has been characterized as an antigen and to be important for virulence mostly by comparison of M. tuberculosis laboratory strains with M. bovis BCG. Its potential as a drug target, as a vaccine candidate, or importantly in diagnosis have been extensively discussed (for a review see, Ref. 48). Our data indicate that the role of ESAT-6 in the pathogenesis of tuberculosis is more complex than expected.
In summary, our data show that many of the well characterized factors such as devR, phoP, and others are equally abundant in our model and most probably do not participate in the attenuation of virulence in this case. Our in-depth proteomics analysis allowed us to question a straightforward role of ESAT-6 in the pathogenesis of tuberculosis. Although it may be an important feature when comparing M. tuberculosis laboratory strains with attenuated BCG strains, there may be other virulence factors of importance expressed by clinical strains. For example, we identified several transcriptional regulators that may play a role in virulence. Our results illustrate the full potential of a proteomics approach to select promising candidate molecules and genes for further characterization using the tools of molecular biology.
Supplementary Material
Acknowledgments
We thank the Proteomic unit (PROBE), University of Bergen, for analytical services.
Footnotes
* This work was supported by the Norwegian Research Council (Project 175141), European Commission 6th framework program (Project 037919), and South Africa-Norway Programme on Research Co-operation (Project 64495).
 This article contains supplemental Figs. 1 and 2 and Files S1–S3.
This article contains supplemental Figs. 1 and 2 and Files S1–S3.
1 The abbreviations used are:
- TB
- tuberculosis
- emPAI
- exponentially modified protein abundance index
- ESAT-6
- early secretory antigenic target, 6 kDa
- PDIM
- phthiocerol dimycocerosate
- PGL
- phenolic glycolipid
- 2CR
- two-component regulatory
- LTQ
- linear quadrupole ion trap
- BCG
- bacillus Calmette-Guérin.
REFERENCES
- 1.Gagneux S., DeRiemer K., Van T., Kato-Maeda M., de Jong B. C., Narayanan S., Nicol M., Niemann S., Kremer K., Gutierrez M. C., Hilty M., Hopewell P. C., Small P. M. (2006) Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dormans J., Burger M., Aguilar D., Hernandez-Pando R., Kremer K., Roholl P., Arend S. M., van Soolingen D. (2004) Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin. Exp. Immunol. 137, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manca C., Tsenova L., Bergtold A., Freeman S., Tovey M., Musser J. M., Barry C. E., 3rd, Freedman V. H., Kaplan G. (2001) Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. U.S.A. 98, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manca C., Reed M. B., Freeman S., Mathema B., Kreiswirth B., Barry C. E., 3rd, Kaplan G. (2004) Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect. Immun. 72, 5511–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahairas G. G., Sabo P. J., Hickey M. J., Singh D. C., Stover C. K. (1996) Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178, 1274–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parish T., Smith D. A., Kendall S., Casali N., Bancroft G. J., Stoker N. G. (2003) Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mougous J. D., Senaratne R. H., Petzold C. J., Jain M., Lee D. H., Schelle M. W., Leavell M. D., Cox J. S., Leary J. A., Riley L. W., Bertozzi C. R. (2006) A sulfated metabolite produced by stf3 negatively regulates the virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 4258–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAdam R. A., Quan S., Smith D. A., Bardarov S., Betts J. C., Cook F. C., Hooker E. U., Lewis A. P., Woollard P., Everett M. J., Lukey P. T., Bancroft G. J., Jacobs, W. R., Duncan K. (2002) Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 9.Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., Kaplan G., Barry C. E., 3rd (2004) A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84–87 [DOI] [PubMed] [Google Scholar]
- 10.Sinsimer D., Huet G., Manca C., Tsenova L., Koo M. S., Kurepina N., Kana B., Mathema B., Marras S. A., Kreiswirth B. N., Guilhot C., Kaplan G. (2008) The phenolic glycolipid of Mycobacterium tuberculosis differentially modulates the early host cytokine response but does not in itself confer hypervirulence. Infect. Immun. 76, 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 12.Tsolaki A. G., Hirsh A. E., DeRiemer K., Enciso J. A., Wong M. Z., Hannan M., Goguet de la Salmoniere Y. O., Aman K., Kato-Maeda M., Small P. M. (2004) Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. U.S.A. 101, 4865–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filliol I., Motiwala A. S., Cavatore M., Qi W., Hazbón M. H., Bobadilla del Valle M., Fyfe J., García-García L., Rastogi N., Sola C., Zozio T., Guerrero M. I., León C. I., Crabtree J., Angiuoli S., Eisenach K. D., Durmaz R., Joloba M. L., Rendón A., Sifuentes-Osornio J., Ponce de León A., Cave M. D., Fleischmann R., Whittam T. S., Alland D. (2006) Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188, 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren R. M., Richardson M., Sampson S. L., van der Spuy G. D., Bourn W., Hauman J. H., Heersma H., Hide W., Beyers N., van Helden P. D. (2001) Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis 81, 291–302 [DOI] [PubMed] [Google Scholar]
- 15.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 16.Hu Q., Noll R. J., Li H., Makarov A., Hardman M., Graham Cooks R. (2005) The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443 [DOI] [PubMed] [Google Scholar]
- 17.Makarov A., Denisov E., Lange O., Horning S. (2006) Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 17, 977–982 [DOI] [PubMed] [Google Scholar]
- 18.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 19.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 20.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 21.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 22.Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D. (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Spuy G. D., Kremer K., Ndabambi S. L., Beyers N., Dunbar R., Marais B. J., van Helden P. D., Warren R. M. (2009) Changing Mycobacterium tuberculosis population highlights clade-specific pathogenic characteristics. Tuberculosis 89, 120–125 [DOI] [PubMed] [Google Scholar]
- 24.Hanekom M., van der Spuy G. D., Streicher E., Ndabambi S. L., McEvoy C. R., Kidd M., Beyers N., Victor T. C., van Helden P. D., Warren R. M. (2007) A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45, 1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Pando R., Orozco H., Arriaga K., Sampieri A., Larriva-Sahd J., Madrid-Marina V. (1997) Analysis of the local kinetics and localization of interleukin-1 alpha, tumour necrosis factor-alpha and transforming growth factor-beta, during the course of experimental pulmonary tuberculosis. Immunology 90, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernández-Pando R., Orozcoe H., Sampieri A., Pavón L., Velasquillo C., Larriva-Sahd J., Alcocer J. M., Madrid M. V. (1996) Correlation between the kinetics of Th1, Th2 cells and pathology in a murine model of experimental pulmonary tuberculosis. Immunology 89, 26–33 [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez-Pando R., Pavön L., Arriaga K., Orozco H., Madrid-Marina V., Rook G. (1997) Pathogenesis of tuberculosis in mice exposed to low and high doses of an environmental mycobacterial saprophyte before infection. Infect. Immun. 65, 3317–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Infante E., Aguilar L. D., Gicquel B., Pando R. H. (2005) Immunogenicity and protective efficacy of the Mycobacterium tuberculosis fadD26 mutant. Clin. Exp. Immunol. 141, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarnagin J. L., Luchsinger D. W. (1980) The use of fluorescein diacetate and ethidium bromide as a stain for evaluating viability of mycobacteria. Stain Technol. 55, 253–258 [DOI] [PubMed] [Google Scholar]
- 30.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 31.Schulze W. X., Mann M. (2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 32.Shi L., Jung Y. J., Tyagi S., Gennaro M. L., North R. J. (2003) Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. U.S.A. 100, 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campuzano J., Aguilar D., Arriaga K., León J. C., Salas-Rangel L. P., González-y-Merchand J., Hernández-Pando R., Espitia C. (2007) The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine 25, 3722–3729 [DOI] [PubMed] [Google Scholar]
- 34.de Souza G. A., Målen H., Søfteland T., Saelensminde G., Prasad S., Jonassen I., Wiker H. G. (2008) High accuracy mass spectrometry analysis as a tool to verify and improve gene annotation using Mycobacterium tuberculosis as an example. BMC Genomics 9, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon S. V., Bottai D., Simeone R., Stinear T. P., Brosch R. (2009) Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. BioEssays 31, 378–388 [DOI] [PubMed] [Google Scholar]
- 36.ten Bokum A. M., Movahedzadeh F., Frita R., Bancroft G. J., Stoker N. G. (2008) The case for hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol. 16, 436–441 [DOI] [PubMed] [Google Scholar]
- 37.Lipsitch M., Moxon E. R. (1997) Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 5, 31–37 [DOI] [PubMed] [Google Scholar]
- 38.Apt A., Kramnik I. (2009) Man and mouse TB: contradictions and solutions. Tuberculosis 89, 195–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. (1999) Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267 [DOI] [PubMed] [Google Scholar]
- 40.Siméone R., Constant P., Guilhot C., Daffé M., Chalut C. (2007) Identification of the missing trans-acting enoyl reductase required for phthiocerol dimycocerosate and phenolglycolipid biosynthesis in Mycobacterium tuberculosis. J. Bacteriol. 189, 4597–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiman D. E., Kaushal D., Ko C., Tyagi S., Manabe Y. C., Schroeder B. G., Fleischmann R. D., Morrison N. E., Converse P. J., Chen P., Bishai W. R. (2004) Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72, 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R., Converse P. J., Ko C., Tyagi S., Morrison N. E., Bishai W. R. (2004) Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52, 25–38 [DOI] [PubMed] [Google Scholar]
- 43.Parish T., Smith D. A., Roberts G., Betts J., Stoker N. G. (2003) The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 44.Malhotra V., Sharma D., Ramanathan V. D., Shakila H., Saini D. K., Chakravorty S., Das T. K., Li Q., Silver R. F., Narayanan P. R., Tyagi J. S. (2004) Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231, 237–245 [DOI] [PubMed] [Google Scholar]
- 45.Kendall S. L., Withers M., Soffair C. N., Moreland N. J., Gurcha S., Sidders B., Frita R., Ten Bokum A., Besra G. S., Lott J. S., Stoker N. G. (2007) A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 65, 684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sørensen A. L., Nagai S., Houen G., Andersen P., Andersen A. B. (1995) Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63, 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis K. N., Liao R., Guinn K. M., Hickey M. J., Smith S., Behr M. A., Sherman D. R. (2003) Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brodin P., Rosenkrands I., Andersen P., Cole S. T., Brosch R. (2004) ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12, 500–508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.