Abstract
IgA nephropathy (IgAN) is the most common primary glomerulonephritis in the world. Aberrantly glycosylated IgA1, with galactose (Gal)-deficient hinge region (HR) O-glycans, plays a pivotal role in the pathogenesis of the disease. It is not known whether the glycosylation defect occurs randomly or preferentially at specific sites. We have described the utility of activated ion-electron capture dissociation (AI-ECD) mass spectrometric analysis of IgA1 O-glycosylation. However, locating and characterizing the entire range of O-glycan attachment sites are analytically challenging due to the clustered serine and threonine residues in the HR of IgA1 heavy chain. To address this problem, we analyzed all glycoforms of the HR glycopeptides of a Gal-deficient IgA1 myeloma protein, mimicking the aberrant IgA1 in patients with IgAN, by use of a combination of IgA-specific proteases + trypsin and AI-ECD Fourier transform ion cyclotron resonance (FT-ICR) tandem mass spectrometry (MS/MS). The IgA-specific proteases provided a variety of IgA1 HR fragments that allowed unambiguous localization of all O-glycosylation sites in the six most abundant glycoforms, including the sites deficient in Gal. Additionally, this protocol was adapted for on-line liquid chromatography (LC)-AI-ECD MS/MS and LC-electron transfer dissociation MS/MS analysis. Our results thus represent a new clinically relevant approach that requires ECD/electron transfer dissociation-type fragmentation to define the molecular events leading to pathogenesis of a chronic kidney disease. Furthermore, this work offers generally applicable principles for the analysis of clustered sites of O-glycosylation.
Glycosylation is one of the most common post-translational modifications of proteins. It is estimated that over half of mammalian proteins are glycosylated. Patients with several autoimmune disorders, chronic inflammatory diseases, and some infectious diseases exhibit abnormal glycosylation of serum immunoglobulins and other glycoproteins (1–5). The biological functions of these modifications in health and disease have become a significant area of interest in biomedical research (6). A subset of these glycoproteins has clustered sites of O-glycosylation with serine- and threonine-rich stretches within the amino acid sequence. Mucins, such as membrane-associated MUC1, are perhaps the best known family of proteins that are heavily O-glycosylated. Their altered expression and aberrant glycosylation have made them potential targets as biomarkers for early detection of cancer (7). Immunoglobulin A1 (IgA1)1 contains both O- and N-glycans (Fig. 1). Aberrant O-glycosylation of IgA1 is involved in the pathogenesis of IgA nephropathy (IgAN) and the closely related Henoch-Schönlein purpura nephritis (1, 8). Interestingly, the aberrantly glycosylated molecules, IgA1 in IgAN and MUC1 in cancer, are recognized by the immune system as neoepitopes as evidenced by formation of specific antibodies (9–11). Mucin-like bacterial surface proteins exhibit similar properties: the molecules have clustered bacterial O-glycans that mediate cellular adhesion, and blocking antibodies target these glycan-containing epitopes (12).
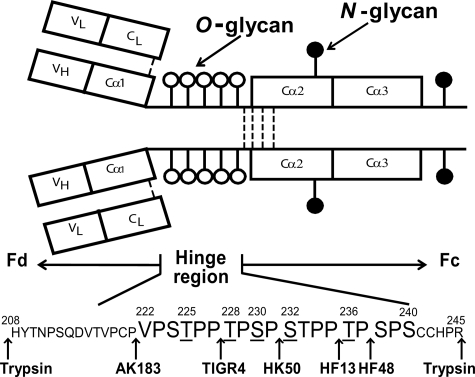
Fig. 1.
IgA1 structural elements. IgA1 has N-linked glycans (filled circles) and O-linked glycans (open circles). The O-glycosylated sites are in the HR between the first and second constant region domains of the heavy chains. The HR is a Pro-rich segment with nine possible sites of O-glycan attachment. Underlined serine and threonine residues are usually glycosylated (31). Arrows show cleavage sites of trypsin and IgA-specific proteases.
An O-glycosylated protein from a single source contains a population of variably O-glycosylated isoforms that show a distinct distribution of microheterogeneity of the O-glycan chains in terms of number, sites of attachment, and composition. Characterizing these clustered sites and understanding how the distributions change under different biological conditions or disease states are an analytical challenge. Enzymatic or chemical release of O-glycans is not selective. The heterogeneity, composition, and quantitative aspects of different O-glycan chains can be assessed and quantified by gas chromatographic and/or mass spectrometric techniques. However, the site-specific information and context of location and composition of adjacent chains are lost. Carbohydrate-specific lectin analysis of O-glycoproteins can provide information on glycan composition and comparative differences between samples, such as those from healthy controls and patients with various disease states. We have successfully demonstrated this in the analysis of IgA1 O-glycans from patients with IgAN versus healthy controls and disease controls (13–15). This included proximal assessment of sites with galactose (Gal)-deficient O-glycans after digests with IgA-specific proteases (8). Several studies have demonstrated the value of mass spectrometry (MS) in identifying Gal-deficient IgA1 in patients with IgAN (16–21), including our work that demonstrated the first direct localization of native sites of O-glycan chains in the hinge region (HR) of IgA1 by use of electron capture dissociation (ECD) (20, 22). ECD and the more recently developed electron transfer dissociation (ETD) have been used to identify sites of O-glycosylation on a variety of proteins (23–26). This includes the analysis of sites of O-glycosylation by on-line LC-ECD/ETD MS/MS methods (23, 26, 27).
IgAN is the most common primary glomerulonephritis worldwide (28) with about 20–40% of patients developing end stage renal failure. It is characterized by mesangial deposits of IgA1-containing immune complexes (28). The distinctive O-glycan chains of IgA1 molecules play a pivotal role in the pathogenesis of IgAN (1, 10, 14–16, 29, 30). IgA1 contains an HR between the first and second heavy chain constant region domains with a high content of Ser, Thr, and Pro. This segment usually has three to five O-glycan chains per HR (31) (see Fig. 1). Aberrantly glycosylated IgA1, deficient in Gal in some of the O-glycans in the HR, in serum is rare in healthy individuals but is present at elevated levels in IgAN patients (13, 15). This distinctive IgA1 is in circulating immune complexes (8, 10, 15) and in the glomerular deposits of IgAN patients (16, 29). The absence of Gal apparently leads to the exposure of neoepitopes, including terminal and sialylated N-acetylgalactosamine (GalNAc) residues (9, 10). These epitopes are recognized by naturally occurring anti-glycan IgG or IgA1 antibodies and, consequently, circulating immune complexes are formed (9, 10, 15) that can deposit in the glomerular mesangia. To identify the pathogenic forms of IgA1, a thorough analysis of O-glycan microheterogeneity, including identification of the attachment sites, will be required.
In this work, we demonstrate the complete analysis of O-glycoform microheterogeneity and site localization of the glycoforms in a naturally Gal-deficient IgA1 (Ale) myeloma protein that mimics the nephritogenic IgA1 in patients with IgAN (8, 9). Reversed phase (RP) LC FT-ICR MS successfully identified 10 distinct IgA1 HR fragments representing >99% of total IgA1. AI-ECD of the six most abundant IgA1 HR glycoforms (>95% of total IgA1) was accomplished with three distinct IgA-specific protease + trypsin digestions, identifying sites of Gal deficiency across four distinct IgA1 O-glycoforms. Based on the success of the ECD fragmentation of these IgA1 HR fragments, we adapted the analysis for on-line LC-MS/MS methods for both ECD and ETD. The variety of IgA1 HR proteolytic fragments provides a practical set of guidelines for the ECD/ETD analysis of clustered sites of O-glycosylation on this and other proteins. These results also provide insight into the order of attachment of the O-glycans in the IgA1 HR.
EXPERIMENTAL PROCEDURES
Sample Preparation
This study was approved by the Institutional Review Board of the University of Alabama at Birmingham. The polymeric form of IgA1 myeloma protein (Ale) was previously isolated from plasma of a patient with multiple myeloma (14). Briefly, the plasma sample was precipitated with ammonium sulfate (50% saturation) and dissolved in phosphate buffer, and IgG and IgM were removed by affinity chromatography with protein G and anti-human IgM antibodies, respectively (32). Next, size-exclusion chromatography on a column of Ultrogel AcA22 (Amersham Biosciences) was used to isolate IgA1. The final purification step included FPLC separation of the polymeric form of IgA1 on a column of Sephacryl 300. The purity of the IgA1 preparation was assessed by SDS-PAGE, Western blotting, and ELISA using polyvalent reagents against IgA and IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) and IgA1-specific and IgA2-specific monoclonal antibodies (15, 32). The polymeric molecular form of the IgA1 (Ale) was assessed by size exclusion chromatography, nonreducing SDS-PAGE, and Western blots developed with anti-IgA antibody. The resultant preparation was polymeric IgA1 that was Gal-deficient in O-glycans, mimicking the glycosylation aberrancy of serum IgA1 in patients with IgAN (8–10, 13–15, 32).
Purified IgA1 protein was treated with an IgA-specific protease from Clostridium ramosum strain AK183 (recombinant in Escherichia coli), Streptococcus pneumoniae strain TIGR4 (recombinant in E. coli), Haemophilus influenzae HK50, Neisseria meningitides strain HF13 (recombinant in E. coli), or N. meningitides strain HF48 followed by trypsin to release IgA1 HR glycopeptides. The digestion with each IgA-specific protease was performed for 24 h at 37 °C in PBS, pH 7.4. Then, trypsin was added and incubated for another 24 h at 37 °C in 100 mm NH4HCO3, pH 8.3. The sequential digestion with AK183 and trypsin released 24-mer HR (Val222–Arg245) glycopeptides, TIGR4 and trypsin generated N-terminal 20-mer (His208–Pro227) and C-terminal 18-mer (Thr228–Arg245) glycopeptides, and HK50 and trypsin produced N-terminal 24-mer (His208–Pro231) and C-terminal 14-mer (Ser232–Arg245) glycopeptides. Furthermore, a shorter IgA1 HR O-glycopeptide, a 16-mer (Val222–Pro237), was prepared by two IgA-specific proteases (AK183 and HF48). Prior to analysis, these digests were treated with 10 milliunits/ml neuraminidase to remove sialic acid residues from the O-glycan chains. After sequential treatment with IgA-specific protease and trypsin, the preparations were reduced with 5 mm dithiothreitol (DTT). Half of each digest was alkylated with 10 mm iodoacetamide. For off-line microscale separation, 25 μg of digested IgA1 was fractionated by RP LC (Agilent 1100 capillary pump) using a 300-μm × 15-cm C18 PepMap column (LC Packings, Sunnyvale, CA) as described previously (22, 33). Eluted IgA1 HR glycopeptides were collected as 5-μl fractions in a 96-well plate by use of an Agilent 1100 microwell plate fraction collector.
On-line LC ESI FT-ICR MS Analyses
On-line LC was performed by use of an Eksigent MicroAS autosampler and 2D LC nanopump (Eksigent, Dublin, CA). Three-hundred nanograms of digested IgA1 was loaded onto a 100-μm-diameter, 11-cm-long column pulled tip packed with Jupiter 5-μm C18 reversed phase beads (Phenomenex). The digests were then eluted with an acetonitrile gradient from 5 to 30% in 0.1% formic acid over 50 min at 650 nl min−1. Linear quadrupole ion trap (LTQ) FT-ICR parameters were set as described previously (22). The mass spectrometer alternated between a full FT MS scan (m/z 400–2,000) and four subsequent MS/MS scans of the four most abundant precursor ions. Survey scans were acquired in the ICR cell with a resolving power of 100,000 at m/z 400. Precursor ions were isolated and subjected to CID in the linear ion trap as confirmation of O-glycopeptide precursor ions. Isolation width was m/z 2. Automatic gain control (AGC) was used to accumulate sufficient precursor ions (target value, 5 × 104 ions; maximum fill period, 250 ms). Dynamic exclusion was enabled with the exclusion window set to 5 ppm with an exclusion time of 120 s after a repeat count of 3 within 60 s. Each analysis of IgA-specific protease + trypsin preparations of IgA1 HR to obtain relative distribution analysis of IgA1 HR O-glycoforms was performed in triplicate with two unique preparations.
AI-ECD FT-ICR MS/MS and ETD LTQ FT MS/MS Analysis
Off line-fractionated IgA1 HR glycopeptides were analyzed by use of a LTQ 7-tesla FT-ICR (LTQ FT) mass spectrometer or an LTQ XL with ETD mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A monolithic silicon microchip-based electrospray interface, TriVersaTM NanoMate (Advion, Ithaca, NY), served as the source for electrospray ionization of the off line-fractionated IgA1 HR O-glycopeptides. AI-ECD FT-ICR MS/MS analysis was performed as described with some modifications (22). The isolation width was m/z 10 with an AGC target value of 2 × 106 and maximal fill period of 3,500 ms. Following transfer to the ICR cell, precursor ion populations were photon-irradiated for 100 ms at 10% (2-Watt) laser power. After 100 ms of photon irradiation, the precursor populations were irradiated with the electrons for 100 ms at 2–3% energy (≈0.5 eV). Each AI-ECD scan was acquired as an FT-ICR broadband mass spectrum (100 < m/z < 3,000) at a mass resolving power of 100,000 at m/z 400. Each displayed spectrum represented a sum of 100 scans. For ETD LTQ XL MS/MS experiments of off-line fractionated IgA1 HR glycopeptides, a range of ETD parameters was tested to optimize fragmentation, and the data were compared with results derived from AI-ECD LTQ FT MS/MS for the same IgA1 HR O-glycoforms. Varied parameters included inclusion of supplemental activation (SA) or not, inclusion of CID of the charge-reduced species (34), and settings for the anion (fluoranthene) injection time (50–500 ms). AGC was set to 1 × 104 for analyte cations and 1.5 × 105 for fluoranthene anions.
On-line ECD/ETD Analysis
On-line LC AI-ECD FT-ICR MS/MS analysis was performed by combining the LC-MS conditions with the off-line AI-ECD FT-ICR MS/MS analysis described above with modifications. The HR O-glycopeptides were eluted with an acetonitrile gradient from 2 to 10% in 0.1% formic acid over 45 min at 650 nl min−1. The isolation width was m/z 7 with an AGC target value of 1 × 106 and maximal fill period of 1,000 ms. Dynamic exclusion parameters were set as described by Creese and Cooper (35).
For on-line LC ETD SA LTQ XL MS/MS analysis, samples were analyzed as reported previously (36) with the exception that the elution gradient was changed to 2–10% over 30 min. AGC was set as above, the analyte injection time was set to 125 ms, and the isolation window was m/z 4 with a repeat count set of 4 within 45 s.
Lectin Western Blot
Samples of IgA1 (Ale) myeloma protein were digested with a single protease at a time (8) and then separated under reducing conditions by SDS-PAGE. The blots were developed with a GalNAc-specific lectin from Helix aspersa (HAA; Sigma-Aldrich) (14).
Data Analysis
All spectra were analyzed by use of the Xcalibur Qual Browser 2.0 software (Thermo Fisher Scientific). Individual IgA1 O-glycopeptides were identified as described previously (22). IgA1 O-glycopeptides species in each LC-MS analysis were identified from the known sequence of the isolated HR glycopeptides, calculated monoisotopic mass of glycopeptides, and the presence of adjacent HR glycopeptides within the high resolution FT-ICR MS spectra. Monoisotopic m/z values for the IgA1 O-glycopeptides were manually tabulated from the raw data files by use of Xcalibur Qual Browser 2.0. The minimal threshold for IgA1 HR glycopeptide identification and relative abundance measurements was a signal to noise ratio of at least 5:1 with >4 isotopic peaks. Identified IgA1 HR O-glycopeptides were checked against theoretical values by use of the GlycoMod tool (http://www.expasy.org). Known IgA1 HR amino acid sequences based on the combination of IgA-specific protease + trypsin digestions performed were inputted with trypsin enzyme and zero missed cleavage sites. Hexose, N-acetylhexosamine, and N-acetylneuraminic acid (NeuAc) monosaccharide residues were all selected as possible (variable) additions to the IgA HR peptides with a mass tolerance of 10 ppm. For alkylated IgA1 HR preparations, cysteine iodoacetamide modification was selected. The relative abundance of each glycopeptide was calculated by use of a label-free quantification method described by Rebecchi et al. (37). After assigning all glycopeptide peaks in the spectrum, the ion chromatogram for each glycopeptide ion was individually extracted for the specific glycopeptide ion species ±0.83 m/z. Then the total ion current from the extracted ion chromatogram of each glycopeptide was divided by the total ion current for all glycopeptides of a given charge state. The relative abundance of each differentially glycosylated IgA1 HR peptide set (IgA-specific protease + trypsin) was calculated as a percentage against total glycopeptides detected. LC-MS replicate runs and differential digestions served as internal controls for assignments of relative abundance. Reported relative abundance measurements for the IgA1 HR glycopeptides in Table I are the average of four independent LC FT-ICR MS analyses of two IgA-specific protease + trypsin digests. For the Thr228–Arg245 C-terminal HR protease-generated fragments, a disaccharide is assumed (>97%; see supplemental Table 1 and supplemental Fig. 4A) at position Thr225 because of the corroborating N-terminal fragment of the same digest and the relative distribution of the TIGR4 + trypsin digestion fragments. Average relative distributions for each observed IgA1 HR O-glycopeptide fragment, observed monoisotopic masses, and mass errors are provided in supplemental Table 1. Theoretical lists of fragmented IgA1 HR peptides by AI-ECD were generated by use of the ProteinProspector MS product tool (http://prospector.ucsf.edu/) with the inclusion of c, z, b, and y ions. Peptide ion fragments of all IgA1 HR AI-ECD FT-ICR MS/MS and ETD LTQ MS/MS spectra were manually assigned to locate sites of O-glycosylation.
Table I. Identified HR O-glycopeptides of IgA1 (Ale) myeloma protein.
A complete list of observed N- and C-terminal fragments, m/z values, and mass errors are provided in supplemental Table 1.
| Glycan structure of identified Val222–Arg245 and Thr228–Arg245O-glycopeptides | Relative abundance ± S.D.a,b |
|---|---|
| % | |
| (GalNAc)6Gal4 | 0.84 ± 0.13 |
| (GalNAc)6Gal3 | 1.19 ± 0.11 |
| (GalNAc)5Gal5 | 5.01 ± 0.92 |
| (GalNAc)5Gal4 | 24.65 ± 4.18 |
| (GalNAc)5Gal4(NeuAc)1 | 0.30 ± 0.14 |
| (GalNAc)5Gal3 | 11.16 ± 1.26 |
| (GalNAc)5Gal2 | 1.09 ± 0.27 |
| (GalNAc)4Gal4 | 34.33 ± 0.80 |
| (GalNAc)4Gal4(NeuAc)1 | 0.57 ± 0.21 |
| (GalNAc)4Gal3 | 18.12 ± 2.82 |
| (GalNAc)4Gal2 | 2.25 ± 0.91 |
| (GalNAc)3Gal3 | 0.49 ± 0.41 |
a Relative abundance of each glycopeptides is expressed as a percentage against the sum of total ion current of glycopeptides detected with the same backbone amino acid sequence, determined similarly to the calculations of Rebecchi et al. (37).
b Relative abundance was calculated from four independent runs based on glycan profiles of Val222–Arg245 and Thr228–Arg245 glycopeptides. Our data showed that Thr225 is almost exclusively present with disaccharide (see supplemental Table 1 and supplemental Fig. 4A).
RESULTS
Proteolytic Release and High Resolution MS Analysis of IgA1 HR Glycopeptides
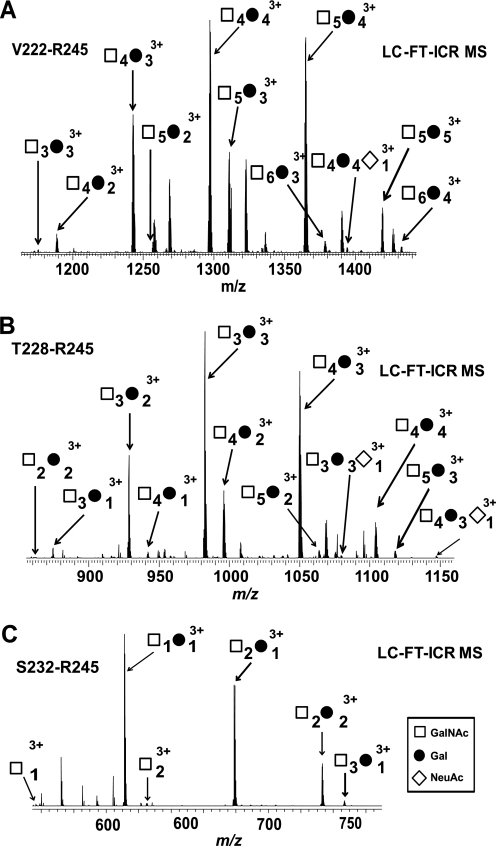
To provide a variety of IgA1 HR O-glycopeptides for analysis by LC-MS and ECD fragmentation, 50 μg of IgA1 (Ale) myeloma protein was digested with three bacterial IgA-specific proteases, one at a time (Fig. 1), followed by trypsin. The digests, with the released series of O-glycopeptides (Table I and supplemental Table 1), were then subjected to on-line LC ESI FT-ICR MS or off-line RP C18 fractionation followed by AI-ECD FT-ICR MS/MS analysis. Three-hundred nanograms of IgA1 was sufficient for on-line LC ESI FT-ICR MS analysis. Fig. 2 shows the FT-ICR mass spectra of IgA1 O-glycopeptides Val222–Arg245 (top), Thr228–Arg245 (middle), and Ser232–Arg245 (bottom) released by AK183, TIGR4, and HK50, respectively, followed by trypsin. The number of O-glycan chains was assigned based on the masses of the amino acid sequence (GalNAc, 203.08 Da; Gal, 162.05 Da; and NeuAc, 291.10 Da) (22). The assignments of hexose and N-acetylhexosamine residues as Gal and GalNAc, respectively, were based on previous glycan compositional analysis (9, 22). Table I lists IgA1 HR O-glycoforms and relative distributions identified by LC-MS calculated as described above. Supplemental Table 1 details the IgA1 HR O-glycopeptides identified in each preparation as well as the abundance of each IgA1 HR O-glycopeptide relative to the other glycoforms with the same amino acid sequence calculated. The TIGR4 and HK50 (+trypsin) digests also generated N-terminal O-glycopeptide HR fragments His208–Pro227 and His208–Pro231 (supplemental Table 1 and supplemental Fig. 1). These results together allowed us to define the number of O-glycopeptides in each individual IgA-specific protease + trypsin digest.
Fig. 2.
IgA1 HR O-glycopeptides generated by differential proteolytic digestions. A combination of IgA-specific proteases and trypsin yielded a series of overlapping C-terminal IgA1 HR O-glycopeptide fragments, Val222–Arg245 (A), Thr228–Arg245 (B), and Ser232–Arg245 (C), that allowed identification of all desialylated IgA1 O-glycoforms in the sample. The distribution of IgA1 O-glycoforms remained the same for the AK183 + trypsin (A) and the TIGR4 + trypsin (B) digests (see supplemental Table 1), indicating that the sites of microheterogeneity are C-terminal to Thr225. The observed N-terminal HR fragments are provided in supplemental Fig. 1.
Locating O-Glycan Attachment Sites
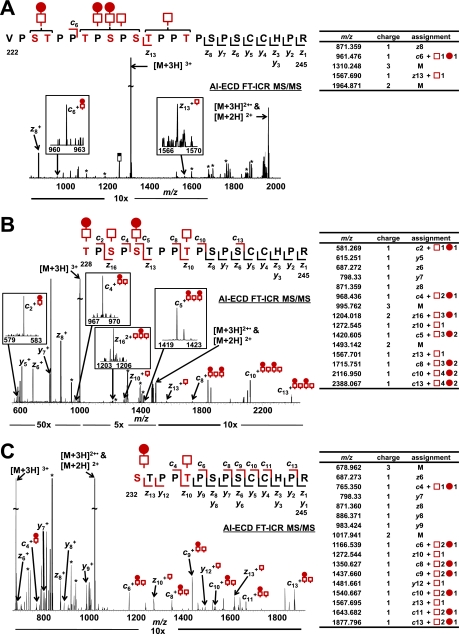
IgA1 HR released by each of the IgA-specific protease + trypsin digestions was isolated using off-line RP C18 LC fractionation (22). Individual HR O-glycoforms were subjected to NanoMate ESI AI-ECD FT-ICR MS/MS to localize all sites of O-glycan attachment in the major O-glycoforms of the IgA1 (Ale) myeloma protein. Fig. 3A displays the AI-ECD spectra for the [Val222–Arg245 + 5 GalNAc + 3 Gal]3+ O-glycopeptide ion generated by AK183 + trypsin digestion. ECD-type c and z fragments within the HR O-glycan cluster allowed location of a single disaccharide N-terminal to the c6 fragment (Ser224 or Thr225), a monosaccharide chain to Thr233 or Thr236, and then, by default, two disaccharide chains and a monosaccharide among Thr228, Ser230, and Ser232. Upon digestion with TIGR4 + trypsin, the five-GalNAc + three-Gal IgA O-glycoform becomes an IgA1 O-glycopeptide (Thr228–Arg245) with four GalNAc and two Gal residues. Fig. 3B shows the AI-ECD MS/MS spectra for [Thr228–Arg245 + 4 GalNAc and 2 Gal]3+ O-glycopeptide ion. Additional ECD fragments were observed within the O-glycan cluster that allowed unambiguous assignment of all four O-glycan chains. From the N terminus, the c2 ion localized a GalNAc-Gal disaccharide to Thr228. The c4 fragment localized a GalNAc monosaccharide to Ser230 followed by the c5 fragment, which assigned a GalNAc-Gal disaccharide to Ser232. The c8 ion indicated that no additional carbohydrates were present; therefore, Thr233 was eliminated as a potential site, and then c10 localized a monosaccharide to Thr236. From the C terminus, four fragments were observed through Ser238 and eliminated this site and Ser240 as potential sites of attachment. The z13 fragment then corroborated the c fragments by showing the addition of only a monosaccharide C-terminal to Thr233 that was already assigned to Thr236. AI-ECD MS/MS of the [Ser232–Arg245 + 2 GalNAc + 1 Gal]3+ O-glycopeptide ion confirmed the assignment of a disaccharide to Ser232 and a monosaccharide to Thr236 (Fig. 3C).
Fig. 3.
AI-ECD fragmentation of (GalNAc)5Gal3 IgA1 HR O-glycoform. AI-ECD FT-ICR MS/MS of the IgA1 HR with five GalNAc and three Gal residues (3+ charge state). The three sets of IgA-specific protease + trypsin fragments provided three distinct O-glycosylated peptides to unambiguously localize all sites of attachment, including two sites of Gal deficiency. Although the AI-ECD MS/MS of the Val222–Arg245 (A) protease-generated fragment did not unambiguously localize all sites of O-glycan attachment, it served to complement and validate the AI-ECD MS/MS data of the Thr228–Arg245 (B) and Ser232–Arg245 (C) fragments. The observed c (above), z, and y (below) fragments for each O-glycopeptide are indicated in the amino acid sequence and adjacent tables. Peaks corresponding to the loss of sugar residues from the precursor ion or charge-reduced species are labeled (*).
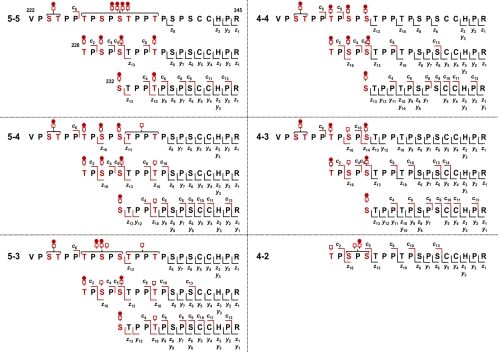
The compiled ECD fragmentation analysis of all three glycopeptides provided unambiguous assignment of all O-glycan chains for the IgA1 O-glycoform with five O-glycan chains, including two glycans that are Gal-deficient. Although the Thr228–Arg245 AI-ECD fragmentation provided the most assignment information, the other two HR O-glycopeptide preparations served to confirm and validate the assignments. A summary of the AI-ECD analyses compiled for the six most abundant IgA1 O-glycoforms is provided in Fig. 4. AI-ECD FT-ICR MS/MS spectra for the six most abundant IgA1 HR O-glycoforms are provided in the supplemental data. The combined AI-ECD analyses and relative abundance analysis (Table I) represent >95% of the total IgA1 in the sample with a defined micro- (amino acid attachment level) and macroheterogeneity (types of glycopeptides in the mixture).
Fig. 4.
AI-ECD MS/MS fragments of six most abundant IgA1 O-glycoforms. The compiled results of AI-ECD fragmentation for the six most abundant IgA1 HR O-glycoforms unambiguously assigned all sites of O-glycosylation in the 16 unique ion species. Observed c, z, and y fragments for each unique O-glycopeptide are indicated above and below the sequences. The microheterogeneity of each of the five sites of O-glycan attachment is also defined in terms of monosaccharide (i.e., Gal-deficient) versus disaccharide composition. The AI-ECD fragmentation of each of the 16 IgA1 O-glycosylated C-terminal fragments indicated that it was dominated by a single ion species with all sites of microheterogeneity C-terminal to Thr225. Thus, the AI-ECD results for the Thr228–Arg245 IgA1 HR protease-generated fragment were the most informative in defining the microheterogeneity. All AI-ECD fragmentation spectra are provided in the supplemental data.
On-line ECD and ETD Analysis
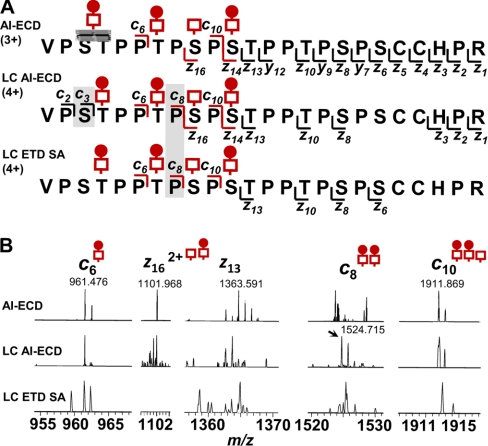
Based on our compiled AI-ECD analysis with different IgA1 HR fragments, we adapted an on-line LC AI-ECD FT-ICR MS/MS method from our LC FT-ICR MS profiling (22) and an LC ECD MS/MS method reported by Creese and Cooper (35) to localize the sites in HR with attached O-glycans. Two LC AI-ECD FT-ICR MS/MS analyses (400 ng/injection) were performed with the Val222–Arg245 and Thr228–Arg245 IgA1 HR proteolytic fragments. To further expand our options for analysis, the same preparations were analyzed by LC ETD on an LTQ XL mass spectrometer. Pilot experiments with an off line-isolated IgA1 HR tryptic fragment, His208–Arg245, confirmed that ETD fragmentation required SA for dissociation as was the case for ECD fragmentation (20) (supplemental Fig. 2). ETD fragmentation of His208–Arg245 did not produce fragments within the clustered sites of O-glycosylation just as we previously saw with ECD of the same IgA1 HR fragment (22) and was recently reported by Wada et al. (38), thereby suggesting that the mechanism of fragmentation was essentially the same. After a similar series of runs to optimize ETD conditions, LC-ETD SA LTQ MS/MS analysis of the IgA-specific protease-trypsin fragments produced fragmentation that was successful in locating the sites of O-glycosylation as was true for AI-ECD MS/MS (supplemental Fig. 3). Fig. 5 shows a comparison between the fragments observed by use of NanoMate ESI AI-ECD, LC AI-ECD, and LC ETD for the Val222–Arg245 + (GalNAc)4Gal3 HR. Under optimized conditions for on-line LC MS/MS analysis, the IgA1 O-glycoform was isolated as a 4+ ion species. More ECD/ETD fragments were observed within the cluster of O-glycan sites compared with off-line preparations of the same peptide. Fig. 5B compares individual ECD/ETD fragments from the off-line and on-line approaches for analyzing these sites of clustered O-glycans and demonstrates the improved fragmentation with the 4+ ion species and considerably less starting material for the analysis (10 μg versus 400 ng).
Fig. 5.
LC-ECD/ETD fragmentation of IgA1 O-glycopeptides. Comparison of the observed on-line LC-ECD/ETD fragmentation for the Val222–Arg245 IgA1 HR + (GalNAc)4Gal3 O-glycopeptide 4+ ion with that of the 3+ charged ion in the off-line fractionated sample is shown (A). The ECD/ETD of the 4+ charged ion species was more successful in distinguishing individual sites of attachment and defining site microheterogeneity because of additional c fragments (c3 and c8, light gray shading in sequence compared to the ambiguous assignment (dark gray) in the off-line AI-ECD of the 3+ ion). When individual fragment ions were examined in the spectra (B), the high resolution LC AI-ECD FT-ICR MS/MS fragments were easier to identify a priori compared with the LC ETD LTQ XL MS/MS fragments. Comparison of several IgA1 HR O-glycoforms selected in data-dependent LC ECD and LC ETD analyses is provided in supplemental Fig. 3.
Defining Overall Microheterogeneity of IgA1 O-Glycans
For most O-glycoforms of IgA1 (Ale) myeloma protein, the microheterogeneity of each site is well defined by our results (Fig. 6). Thr225 was predominantly modified with a GalNAc-Gal disaccharide. The Thr228 O-glycan is predominantly a disaccharide with the exception of the Thr228–Arg245 + (GalNAc)3Gal1 glycoform (inferred to be (GalNAc)4Gal2) with a Gal-deficient site at Thr228. Ser232 contains also predominantly a disaccharide. Although we were not able to unambiguously assign the disaccharide and a monosaccharide between Ser230 and Ser232 in the (GalNAc)4Gal2 O-glycoform, the complete assignment of the (GalNAc)4Gal3 O-glycoform would support the same conclusion. Ser230 and Thr236 are clearly the dominant sites of Gal deficiency. The two glycoforms with an unambiguous monosaccharide assigned at Ser230 represent >29% of the total IgA1 HR within the sample. The three IgA-specific protease + trypsin digests indicate that ∼35% of the IgA1 HR in the sample has a monosaccharide at Thr236. Lectin Western blot analysis with GalNAc-specific lectin (HAA) of IgA1 after digestion with IgA-specific proteases supports the interpretation that these two sites are the dominant sites of Gal deficiency as deduced from the split HAA reactivity between the Fc and Fd fragments for the HK50 and HF13 digestions (Fig. 6B). These two sites together represented >50% of Gal-deficient O-glycosylated IgA1 HR species within the sample. In summary, these results identify >99% of the total IgA1 O-glycoforms in the sample (Table I and supplemental Table 1) and define the site-specific microheterogeneity of >95% (six most abundant glycoforms, five sites of glycosylation) of the total IgA1 (Ale) myeloma protein. For the six most abundant glycoforms, our analysis provides the complete amino acid context of each O-glycoform.
Fig. 6.
Summary of analysis of O-glycosylation of IgA1 (Ale) myeloma protein. The combined LC FT-ICR MS and AI-ECD MS/MS data provided comprehensive analysis of all sites of attachment and single site and overall O-glycan microheterogeneity distribution of IgA1 HR O-glycoforms (A). These results, which provided a high level of detail to define the composition of >95% of IgA in the sample, agreed with those of HAA lectin (GalNAc-specific lectin) Western blot analysis after the IgA1 sample was digested with a panel of IgA-specific proteases (B). Positions of Fc (larger) and Fd (smaller) fragments are marked on the side of the blot.
DISCUSSION
IgA1 myeloma proteins frequently exhibit a high abundance of Gal-deficient O-glycans similar to those on nephritogenic IgA1 in patients with IgAN (8–10, 13–15), and because they are readily available in large amounts, they serve as convenient model proteins. Our combined ECD localization and accurate mass profiles of IgA1 HR O-glycan microheterogeneity provide a full scale picture of the range of IgA1 O-glycoforms isolated from a single source (polymeric IgA1 myeloma protein) (20, 22). In our previous experiments, we observed certain IgA1 O-glycoforms for which we and others (38) could not unambiguously localize all sites of O-glycosylation especially in the more heavily glycosylated (>4 O-glycans) species. The suppression of ECD fragmentation between amino acids within the clustered O-glycan region hampered complete characterization of all O-glycoforms within the sample. This suppressive effect has been observed for N-glycopeptides as well (39). To identify which IgA1 glycoforms are involved in the pathogenesis of IgAN, all sites with Gal deficiency need to be identified. Our goal with this model protein was to define the macro- and microheterogeneity for each site of O-glycosylation across the majority of, if not all, O-glycoforms in the sample toward the long term goal of accomplishing the same with clinical samples. We hypothesized that a shorter IgA1 HR that still retained the C-terminal Arg245 and nearby His243 would result in more extensive fragmentation within the cluster of O-glycosylation sites. Our ECD/ETD analyses of the six most abundant IgA1 O-glycoforms confirmed this hypothesis. Table I and Fig. 6 summarize our combined analyses to provide a comprehensive picture of the O-glycan microheterogeneity within the IgA1 (Ale) myeloma protein. Twenty-six sites of O-glycosylation across six unique IgA1 HR glycopeptide ion species were unambiguously localized. Four other low abundance nonsialylated O-glycoforms were also identified for both the Val222–Arg245 and Thr228–Arg245 HR fragments (Table I). The nearly identical distribution of IgA1 HR O-glycoforms between these two HR preparations (supplemental Table 1) indicated that the major sites of heterogeneity were C-terminal to Thr225. The dominant ion species for the complementary N-terminal fragments (His208–Pro227 and His208–Pro231; supplemental Table 1) were in agreement with the C-terminal fragment distributions that would assign a disaccharide at Thr225 for the majority of species in the sample (>97%).
In contrast, two minor His208–Pro227 peptide ions were observed (supplemental Fig. 1 and Table I), one with a single GalNAc and the other as an unmodified peptide. This finding suggested a site of Gal deficiency at Thr225; however, based on the similar distribution of Val222–Arg245 and Thr228–Arg245 fragments, we reasoned that it would be only a small contribution to the total population of Gal-deficient sites as supported by lectin Western blot analysis (Fig. 6B). In all LC-MS analyses, there was no indication of loss of GalNAc or Gal residues in the ionization process. Comparison of total ion current values for these N-terminal fragments with those for their C-terminal counterparts suggests that the ionization efficiencies for the N-terminal fragments may be considerably higher, thus increasing the sensitivity for these low abundance glycoforms. Additionally, three minor His208–Pro231 glycopeptides with (GalNAc)3Gal1, (GalNAc)2Gal2, and (GalNAc)2Gal1 attached were also observed. The results from both N-terminal HR peptides support the existence of a small population of underglycosylated and Gal-deficient O-glycoforms N-terminal to Ser232. This finding means that the dominant IgA1 O-glycoforms in the C-terminal HR fragments could include low abundance mixtures of equally glycosylated isobaric forms. We have previously demonstrated that AI-ECD of IgA1 HR distinguished between a single population and an isobaric mixture of differentially glycosylated HR peptides (20). However, fragmentation by use of ECD is less efficient than the more traditional collision-induced dissociation in the conversion of parent ions to fragments (∼30 versus >70% conversion) (40). As such, a small subpopulation of isobaric IgA1 HR O-glycoforms with sites of Gal deficiency as indicated by the N-terminal fragments would not be observed in the presence of a dominant single population. Our relative quantitation of the IgA1 HR O-glycoform distributions was generated from three independent IgA-specific protease + trypsin digestions. The distributions observed for each HR protease-generated fragment corroborate and provide an internal check for the assigned relative distributions. Heterogeneity of clustered O-glycans requires definition of microheterogeneity of individual amino acid attachment sites and of the adjacent O-glycans to provide a full scope of the distribution. Cross-validation between differentially digested HR fragments makes this possible.
We propose that the distribution of IgA1 (Ale) myeloma protein O-glycosylation is dominated by a collection of IgA1 O-glycoforms that may originate from semiordered carbohydrate additions that proceed from Thr225 to Thr236. There also exists a smaller subpopulation of IgA1 O-glycoforms that does not follow this semiordered carbohydrate addition, giving rise to differentially O-glycosylated isobaric species. Wandall et al. (41) reported that the lectin domains of different GalNAc-transferases can recognize the same substrate. This could result in different sites or order of GalNAc attachment catalyzed by several GalNAc-transferases. Alternatively, there may be a low level natural occurrence of Thr225 being “skipped” by the GalNAc-transferase enzyme. Either explanation is plausible.
The distribution of HR glycoforms for this IgA1 myeloma protein centers around the glycopeptides with (GalNAc)4Gal3, (GalNAc)4Gal4, and (GalNAc)5Gal4, similar to our previously reported IgA1 (Mce) myeloma protein and that of serum nephritogenic IgA1 (17, 18). The assigned sites of glycosylation suggest a semiordered addition proceeding from the Thr225 site. We propose that the (GalNAc)4Gal4 IgA1 HR species is the product of the addition of a Gal residue to the GalNAc monosaccharide at Ser230 of the (GalNAc)4Gal3 IgA O-glycoform. Alternatively, but to a lesser extent, a GalNAc-transferase can add a single GalNAc monosaccharide to Thr236, giving rise to the (GalNAc)5Gal3 IgA1 O-glycoform. Working in the opposite direction, the (GalNAc)4Gal3 IgA1 O-glycoform is the product of the addition of a Gal to the Thr228 of the (GalNAc)4Gal2 IgA1 O-glycoform or, to a lesser extent, the addition of a GalNAc residue to Ser230 to the low abundance (GalNAc)3Gal3. Based on the relative abundance, the former is the more likely route, and the (GalNAc)4Gal2 O-glycoform is alternatively the substrate for addition of a GalNAc residue to Thr236. Therefore, the GalNAc-transferase is proceeding “ahead” of the Gal-transferase, adding GalNAc residues to Thr225 first and then Thr228 followed by Ser230 and Ser232 in some order, then by Thr236, and finally by a sixth undetermined site. The Gal-transferase in tandem appears to add Gal residues to the GalNAc residues first to Thr225 followed by Ser232 at a nearly equal rate but then lags behind slightly for the attachment at Thr228 and even more so at Ser230 and Thr236, thereby accounting for the predominant sites of Gal deficiency.
Application of ECD/ETD to Clustered Sites of O-Glycosylation
Our collection of IgA1 HR fragments (Table II) provides a unique combination of fragments to consider fundamental aspects of ECD/ETD fragmentation techniques analyzing clustered O-glycans. In our previous (20, 22) and current analyses of IgA1 HR proteolytic fragments, supplemental activation was required to produce ECD/ETD fragmentation sufficient to localize sites of attachment. All IgA1 HR glycopeptides fragmented were either 3+ or 4+charged. However, we could unambiguously assign sites of O-glycosylation for only the IgA1 HR peptides that contained the C-terminal Arg245 and the adjacent His243. ECD fragmentation of the Val222–Pro237 IgA1 HR peptide (3+) resulted in a dominant loss of sugars and the absence of any c- or z-type fragments (22). Given that the electron capture/transfer occurs at the sites of proton attachment (42), amino acid proton affinities (43) for the various IgA1 HR fragments could provide insight into the varied success in fragmentation. For the 3+charged Val222–Pro237 fragments, the charge would be assigned to the N-terminal amino group and then the two sites of adjacent prolines within the O-glycan cluster. These charged sites would be the logical sites of electron capture or transfer and may explain the resultant dominance of fragments with loss of hexose and N-acetylhexosamine. The two longest HR fragments (Val216–Leu246 and His208–Arg245) contain the C-terminal Arg245 and adjacent His243 residues. In our previous analysis, these O-glycopeptides were observed as 3+ and 4+ charged ion species that were successful in producing ECD/ETD-type fragments that localized most (but not all) sites of O-glycosylation. Based on proton affinities, the associated charged sites would be assigned to the N-terminal amino group and then the Arg245 and His243 side chains, unlike the Val222–Pro237 fragment. Still, there were several IgA1 O-glycoforms, notably the more heavily glycosylated forms with four and five chains, for which the fragments were either of low abundance or absent within the clustered region of O-glycans (Thr225–Thr236). In our current results, the combined IgA-specific protease + trypsin digestions generated IgA1 HR fragments with the C-terminal Arg245 and His243 residues and a varied N terminus, depending on the IgA-specific protease. Loss of carbohydrate residues was observed for these fragments (Fig. 3 and supplemental Figs. 2 and 4). However, this loss of carbohydrate residues was not the only fragment observed as with the previous Val222-Pro237 fragment (22). There were a variety of c and z fragments that allowed unambiguous assignment of all sites reported above. In contrast, AI-ECD MS/MS of the N-terminal IgA1 O-glycopeptides (His208–Pro227 and His208–Pro231; supplemental Fig. 4) with an N-terminal histidine and one or two pairs of adjacent prolines resulted in a series of ambiguous c, z, and y fragments with and without carbohydrate residues.
Table II. IgA1 HR fragments analyzed by ECD/ETD fragmentation.
§ The residues in red are sites of O-glycan attachment. The underlined residues are the likely basic sites of proton attachment.
† ECD MS/MS spectra of the IgA1 HR N-terminal peptides are provided in supplemental Fig. 4.
The success of ECD/ETD peptide fragmentation with higher charge state peptide ions is well established (42, 44). Broader studies on peptide and glycopeptide ETD fragmentation have indicated that peptide charge density (residues per charge) plays a significant role in the amount of backbone fragmentation (45, 46). The difference in observed fragments for the 3+ and 4+charged Val222–Arg245 IgA1 HR peptides demonstrates this principle for improving ETD/ECD fragmentation in the analysis of clustered sites of O-glycans as well. Clustered O-glycans significantly increase the m/z of precursor cations, thus lowering the charge density. The consistent requirement for supplemental activation in our current and previous studies (20, 22) of IgA1 O-glycopeptides corroborates broader observations on its use to disrupt non-covalent interactions after ECD/ETD and increase the fragmentation efficiency of higher charge state precursor cations (45, 47). Several groups have also reported the use of different proteases to change the relative location of the charge within peptides to optimize ECD/ETD peptide fragmentation (44, 48). There have also been studies on the chemical modification of side chains or C termini to produce higher charge state peptides (49). The IgA1 HR contains two cysteine residues that provide the opportunity to increase charge density in future studies. In our analysis, the proximity of the sites of proton attachment and subsequent electron capture/transfer seems to also play a role in the success of locating clustered sites of O-glycosylation and not just an indiscriminate charge density. This also has been reported in broader studies for doubly charged precursor peptide cations (50). However, the bias is said to decrease with higher charge states. The IgA-specific protease + trypsin fragments do produce lower m/z (i.e., higher charge density) glycopeptides compared with the Val216–Leu246 and His208–Arg245 fragments. Although the Val222–Pro237 fragments would have comparable charge density, the ECD did not produce fragments sufficient to localize any sites of glycan attachment (22). Our collection of IgA1 HR clustered O-glycopeptides indicates that if the electron capture/transfer is too close to the attached glycans (Val222–Pro237) there may be a resulting dominant loss of carbohydrates. If it occurs too far from the attached glycans (Val216–Leu246 and His208–Arg245), there may be insufficient fragmentation for all glycoforms. A blend of the two, with two or three different N termini, provides a compiled unambiguous assignment of all sites of attachment. These empirical conclusions could be applied to other proteins with similar clustered sites. The IgA1 HR has fortuitous basic residues within 10 amino acids of the sites of interest. For other proteins, the amino acid sequence may not be as accommodating. Interestingly, the ECD/ETD fragments that were crucial in defining the IgA1 O-glycan assignments and heterogeneity of individual sites came from the varied N-terminal c fragments between Thr228 and Thr236. This finding suggests that a distal site for electron attachment is not the only requirement. This hypothesis is supported by our previous results with the trypsin fragment (His203–Arg245). The difference in observed fragments for the 3+ and 4+charged Val222–Arg245 IgA1 HR peptides compared with the absence of backbone fragmentation in the Val222–Pro237 glycopeptide provides support for the importance of charge density and of basic residues relative to the clustered sites of interest. Based on proton affinities, the additional charge for the 4+charged Val222–Arg245 ion species could be assigned to either of the two adjacent Pro residues within the O-glycan cluster. This charged ion species would be similar to that of the Val222–Pro237 fragment with the difference being the C-terminal Arg245 and His243. This raises the question of whether a greater portion of electron capture could occur at one site (Arg245 or His243) over another (adjacent Pro residues) to limit the loss of carbohydrate residues but still allow additional c fragments for the 4+ charged Val222–Arg245 fragment.
Within the context of IgAN, locating sites of IgA1 O-glycans with Gal deficiency that lead to formation of IgA1-containing circulating immune complexes requires isolation of a select population of IgA1 molecules from IgAN patients. The IgA1 in IgA1-containing immune complexes is predominantly polymeric (10), whereas monomeric IgA1 comprises >95% of total serum IgA1. From a 1–2-ml serum sample, one can usually isolate 2–4 mg of IgA1, but only 5–10 μg of IgA1 would be within immune complexes2; this small fraction represents the molecules of interest for analysis of O-glycan composition and site localization. As such, off-line preparations of IgA1 for AI-ECD MS/MS characterization that require 10–25 μg of digested IgA1 are not practical for analysis of clinical samples. Our results for on-line ECD/ETD MS/MS analysis demonstrate that individual O-glycoforms can be successfully analyzed on the LC time scale. In a side-by-side comparison (Fig. 5), the O-glycopeptide fragments were recognized by mass and isotope patterns especially with on-line LC AI-ECD FT-ICR MS/MS. For LC ETD SA LTQ XL MS/MS, the lower resolution tandem MS scans rendered assignment of the distinguishing fragments difficult. We presume that a similar analysis with a high resolution LTQ Orbitrap would improve these results as others have shown for O-glycosylation (23). We also observed a greater frequency of hydrogen migration in the ETD with SA. Our AI-ECD method included ion activation by low power infrared multiphoton dissociation prior to electron irradiation (22) and that may explain the observed differences (51, 52). Comparison of on-line ECD fragmentation with ETD fragmentation of the IgA-specific protease-trypsin HR segments showed that fragment ions outside the cluster of O-glycan attachment sites were readily identified in all glycoforms as was observed with the His208–Arg245 segment (supplemental Fig. 2). Within the clustered region, ECD/ETD fragmentation of the IgA-specific protease trypsin fragments was more successful than that of the longer tryptic fragments at defining the sites of disaccharide composition and sites that were Gal-deficient. Several more z and even y fragments were observed in the NanoMate ESI AI-ECD MS/MS analysis for the Ser240–Arg245 region of the HR. This finding is most likely attributable to the higher number of scans (n = 80–100) due to using larger amounts of starting material and an off-line preparation. Results for on-line AI-ECD and ETD with SA were derived from seven or fewer scans.
Implications for IgAN
IgA1 (Ale) and other IgA1 myeloma proteins mimic the Gal-deficient IgA1 in the serum of patients with IgAN as we have demonstrated in previous studies (8–10, 13–15). This IgA1 (Ale) myeloma protein is polymeric and Gal-deficient, reacts with glycan-specific antibodies from sera of IgAN patients, and forms immune complexes that can activate cultured mesangial cells (8, 9).2 However, unlike the Gal-deficient IgA1 in patients with IgAN, the O-glycans of the IgA1 (Ale) myeloma protein is minimally sialylated. Our work lays the foundation for analysis of this nephritogenic IgA1 to define the frequency of the various Gal-deficient glycans and their sites of attachment on the heavy chain. There likely are different sets or classes of Gal-deficient O-glycan-dependent epitopes. These variations may reflect an aberrant activity of the specific glycosyltransferases. However the Gal-deficient species arise, a combined use of IgA-specific protease + trypsin will define the O-glycan-dependent epitope recognized by circulating antibodies that subsequently lead to formation of circulating immune complexes that induce renal injury. Variations in the composition or location of the Gal-deficient glycans may account for differences in the clinical severity of the disease.
Conclusions
Characterization of the molecular details of heterogeneity of the Gal-deficient O-linked glycans on serum IgA1 is vital for understanding the pathogenesis of IgAN. Each IgA1 O-glycoform represents a unique protein. In the case of IgAN, some forms are pathogenic, whereas others may not be. Comprehensive analysis of this post-translational modification requires targeted proteomics preparation engineering based on the amino acid sequence of a given protein and knowledge of how it will fragment. Our analysis of an IgA1 myeloma protein demonstrates that ECD/ETD fragmentation combined with IgA-specific protease digestion can define the distribution and specific sites of Gal deficiency for the majority of the O-glycoforms within a single sample of IgA1. Moreover, this analysis can be accomplished in an on-line LC-MS format. This work represents a significant step forward in the analysis of clustered sites of O-glycosylation. Although specific for the IgA1 HR, there is a commonality of Ser/Thr- and Pro-rich sequences in many of these clustered sites. This study lays the frame-work for successfully defining the sites of attachment, single-site and overall microheterogeneity, and O-glycoform distribution of clustered sites of O-glycosylation, such as those in immunoglobulins, mucins, and bacterial cell-surface proteins. At the same time, these results provide a means to thoroughly characterize aberrant IgA1 O-glycans that contribute to the pathology of the most common form of glomerulonephritis in the world.
Supplementary Material
Acknowledgments
We appreciate Rhubell Brown for assistance in the purification of IgA1 (Ale) myeloma protein and Greg Bowersock for technical assistance with the LC ETD analysis.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants DK077279, DK078244, DK080301, DK075868, DK071802, DK082753, DK083663, and RR17261.
 This article contains supplemental Figs. 1–4 and Table 1. All assigned MS/MS spectra are also provided.
This article contains supplemental Figs. 1–4 and Table 1. All assigned MS/MS spectra are also provided.
2 J. Novak, unpublished data.
1 The abbreviations used are:
- IgA1
- immunoglobulin A1
- IgAN
- IgA nephropathy
- HR
- hinge region
- ECD
- electron capture dissociation
- AI
- activated ion
- ETD
- electron transfer dissociation
- SA
- supplemental activation
- HAA
- GalNAc-specific lectin from Helix aspersa
- RP
- reversed phase
- AGC
- automatic gain control
- LTQ
- linear quadrupole ion trap.
REFERENCES
- 1.Mestecky J., Tomana M., Crowley-Nowick P. A., Moldoveanu Z., Julian B. A., Jackson S. (1993) Defective galactosylation and clearance of IgA1 molecules as a possible etiopathogenic factor in IgA nephropathy. Contrib. Nephrol. 104, 172–182 [DOI] [PubMed] [Google Scholar]
- 2.Moore J. S., Wu X., Kulhavy R., Tomana M., Novak J., Moldoveanu Z., Brown R., Goepfert P. A., Mestecky J. (2005) Increased levels of galactose-deficient IgG in sera of HIV-1-infected individuals. AIDS 19, 381–389 [DOI] [PubMed] [Google Scholar]
- 3.Rademacher T. W., Williams P., Dwek R. A. (1994) Agalactosyl glycoforms of IgG autoantibodies are pathogenic. Proc. Natl. Acad. Sci. U.S.A. 91, 6123–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springer G. F. (1997) Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J. Mol. Med. 75, 594–602 [DOI] [PubMed] [Google Scholar]
- 5.Troelsen L. N., Garred P., Madsen H. O., Jacobsen S. (2007) Genetically determined high serum levels of mannose-binding lectin and agalactosyl IgG are associated with ischemic heart disease in rheumatoid arthritis. Arthritis Rheum. 56, 21–29 [DOI] [PubMed] [Google Scholar]
- 6.Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 7.Storr S. J., Royle L., Chapman C. J., Hamid U. M., Robertson J. F., Murray A., Dwek R. A., Rudd P. M. (2008) The O-linked glycosylation of secretory/shed MUC1 from an advanced breast cancer patient's serum. Glycobiology 18, 456–462 [DOI] [PubMed] [Google Scholar]
- 8.Novak J., Moldoveanu Z., Renfrow M. B., Yanagihara T., Suzuki H., Raska M., Hall S., Brown R., Huang W. Q., Goepfert A., Kilian M., Poulsen K., Tomana M., Wyatt R. J., Julian B. A., Mestecky J. (2007) IgA nephropathy and Henoch-Schoenlein purpura nephritis: aberrant glycosylation of IgA1, formation of IgA1-containing immune complexes, and activation of mesangial cells. Contrib. Nephrol. 157, 134–138 [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H., Fan R., Zhang Z., Brown R., Hall S., Julian B. A., Chatham W. W., Suzuki Y., Wyatt R. J., Moldoveanu Z., Lee J. Y., Robinson J., Tomana M., Tomino Y., Mestecky J., Novak J. (2009) Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J. Clin. Investig. 119, 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomana M., Novak J., Julian B. A., Matousovic K., Konecny K., Mestecky J. (1999) Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J. Clin. Investig. 104, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlad A. M., Kettel J. C., Alajez N. M., Carlos C. A., Finn O. J. (2004) MUC1 immunobiology: from discovery to clinical applications. Adv. Immunol. 82, 249–293 [DOI] [PubMed] [Google Scholar]
- 12.Stephenson A. E., Wu H., Novak J., Tomana M., Mintz K., Fives-Taylor P. (2002) The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43, 147–157 [DOI] [PubMed] [Google Scholar]
- 13.Moldoveanu Z., Wyatt R. J., Lee J. Y., Tomana M., Julian B. A., Mestecky J., Huang W. Q., Anreddy S. R., Hall S., Hastings M. C., Lau K. K., Cook W. J., Novak J. (2007) Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 71, 1148–1154 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H., Moldoveanu Z., Hall S., Brown R., Vu H. L., Novak L., Julian B. A., Tomana M., Wyatt R. J., Edberg J. C., Alarcón G. S., Kimberly R. P., Tomino Y., Mestecky J., Novak J. (2008) IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J. Clin. Investig. 118, 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomana M., Matousovic K., Julian B. A., Radl J., Konecny K., Mestecky J. (1997) Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int. 52, 509–516 [DOI] [PubMed] [Google Scholar]
- 16.Hiki Y., Odani H., Takahashi M., Yasuda Y., Nishimoto A., Iwase H., Shinzato T., Kobayashi Y., Maeda K. (2001) Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int. 59, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 17.Hiki Y., Tanaka A., Kokubo T., Iwase H., Nishikido J., Hotta K., Kobayashi Y. (1998) Analyses of IgA1 hinge glycopeptides in IgA nephropathy by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Am. Soc. Nephrol. 9, 577–582 [DOI] [PubMed] [Google Scholar]
- 18.Novak J., Tomana M., Kilian M., Coward L., Kulhavy R., Barnes S., Mestecky J. (2000) Heterogeneity of O-glycosylation in the hinge region of human IgA1. Mol. Immunol. 37, 1047–1056 [DOI] [PubMed] [Google Scholar]
- 19.Odani H., Hiki Y., Takahashi M., Nishimoto A., Yasuda Y., Iwase H., Shinzato T., Maeda K. (2000) Direct evidence for decreased sialylation and galactosylation of human serum IgA1 Fc O-glycosylated hinge peptides in IgA nephropathy by mass spectrometry. Biochem. Biophys. Res. Commun. 271, 268–274 [DOI] [PubMed] [Google Scholar]
- 20.Renfrow M. B., Cooper H. J., Tomana M., Kulhavy R., Hiki Y., Toma K., Emmett M. R., Mestecky J., Marshall A. G., Novak J. (2005) Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation Fourier transform-ion cyclotron resonance mass spectrometry. J. Biol. Chem. 280, 19136–19145 [DOI] [PubMed] [Google Scholar]
- 21.Tarelli E. (2007) Resistance to deglycosylation by ammonia of IgA1 O-glycopeptides: implications for the β-elimination of O-glycans linked to serine and threonine. Carbohydr. Res. 342, 2322–2325 [DOI] [PubMed] [Google Scholar]
- 22.Renfrow M. B., Mackay C. L., Chalmers M. J., Julian B. A., Mestecky J., Kilian M., Poulsen K., Emmett M. R., Marshall A. G., Novak J. (2007) Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal. Bioanal. Chem. 389, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 23.Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 25.Mirgorodskaya E., Roepstorff P., Zubarev R. A. (1999) Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 71, 4431–4436 [DOI] [PubMed] [Google Scholar]
- 26.Sihlbom C., van Dijk Härd I., Lidell M. E., Noll T., Hansson G. C., Bäckström M. (2009) Localization of O-glycans in MUC1 glycoproteins using electron-capture dissociation fragmentation mass spectrometry. Glycobiology 19, 375–381 [DOI] [PubMed] [Google Scholar]
- 27.Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 28.Julian B. A., Waldo F. B., Rifai A., Mestecky J. (1988) IgA nephropathy, the most common glomerulonephritis worldwide. A neglected disease in the United States? Am. J. Med. 84, 129–132 [DOI] [PubMed] [Google Scholar]
- 29.Allen A. C., Bailey E. M., Brenchley P. E., Buck K. S., Barratt J., Feehally J. (2001) Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: observation in three patients. Kidney Int. 60, 969–973 [DOI] [PubMed] [Google Scholar]
- 30.Novak J., Julian B. A., Tomana M., Mesteck J. (2001) Progress in molecular and genetic studies of IgA nephropathy. J. Clin. Immunol. 21, 310–327 [DOI] [PubMed] [Google Scholar]
- 31.Mattu T. S., Pleass R. J., Willis A. C., Kilian M., Wormald M. R., Lellouch A. C., Rudd P. M., Woof J. M., Dwek R. A. (1998) The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J. Biol. Chem. 273, 2260–2272 [DOI] [PubMed] [Google Scholar]
- 32.Moore J. S., Kulhavy R., Tomana M., Moldoveanu Z., Suzuki H., Brown R., Hall S., Kilian M., Poulsen K., Mestecky J., Julian B. A., Novak J. (2007) Reactivities of N-acetylgalactosamine-specific lectins with human IgA1 proteins. Mol. Immunol. 44, 2598–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan B., Addona T. A., Carr S. A. (2004) Selective detection of glycopeptides on ion trap mass spectrometers. Anal. Chem. 76, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 34.Wu S. L., Hühmer A. F., Hao Z., Karger B. L. (2007) On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J. Proteome Res. 6, 4230–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creese A. J., Cooper H. J. (2007) Liquid chromatography electron capture dissociation tandem mass spectrometry (LC-ECD-MS/MS) versus liquid chromatography collision-induced dissociation tandem mass spectrometry (LC-CID-MS/MS) for the identification of proteins. J. Am. Soc. Mass Spectrom. 18, 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam M. M., Wallin R., Wynn R. M., Conway M., Fujii H., Mobley J. A., Chuang D. T., Hutson S. M. (2007) A novel branched-chain amino acid metabolon. Protein-protein interactions in a supramolecular complex. J. Biol. Chem. 282, 11893–11903 [DOI] [PubMed] [Google Scholar]
- 37.Rebecchi K. R., Wenke J. L., Go E. P., Desaire H. (2009) Label-free quantitation: a new glycoproteomics approach. J. Am. Soc. Mass Spectrom. 20, 1048–1059 [DOI] [PubMed] [Google Scholar]
- 38.Wada Y., Tajiri M., Ohshima S. (2010) Quantitation of saccharide compositions of O-glycans by mass spectrometry of glycopeptides and its application to rheumatoid arthritis. J. Proteome Res. 9, 1367–1373 [DOI] [PubMed] [Google Scholar]
- 39.Håkansson K., Cooper H. J., Emmett M. R., Costello C. E., Marshall A. G., Nilsson C. L. (2001) Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal. Chem. 73, 4530–4536 [DOI] [PubMed] [Google Scholar]
- 40.McFarland M. A., Chalmers M. J., Quinn J. P., Hendrickson C. L., Marshall A. G. (2005) Evaluation and optimization of electron capture dissociation efficiency in Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 1060–1066 [DOI] [PubMed] [Google Scholar]
- 41.Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., Kato K., Irimura T., Suryanarayanan G., Hollingsworth M. A., Clausen H. (2007) The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology 17, 374–387 [DOI] [PubMed] [Google Scholar]
- 42.Iavarone A. T., Paech K., Williams E. R. (2004) Effects of charge state and cationizing agent on the electron capture dissociation of a peptide. Anal. Chem. 76, 2231–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison A. G. (1997) The gas-phase basicities and proton affinities of amino acids and peptides. Mass Spectrom. Rev. 16, 201–217 [Google Scholar]
- 44.Kjeldsen F., Giessing A. M., Ingrell C. R., Jensen O. N. (2007) Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal. Chem. 79, 9243–9252 [DOI] [PubMed] [Google Scholar]
- 45.Darula Z., Medzihradszky K. F. (2009) Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol. Cell. Proteomics 8, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 47.Horn D. M., Ge Y., McLafferty F. W. (2000) Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. Anal. Chem. 72, 4778–4784 [DOI] [PubMed] [Google Scholar]
- 48.Hennrich M. L., Boersema P. J., van den Toorn H., Mischerikow N., Heck A. J., Mohammed S. (2009) Effect of chemical modifications on peptide fragmentation behavior upon electron transfer induced dissociation. Anal. Chem. 81, 7814–7822 [DOI] [PubMed] [Google Scholar]
- 49.Vasicek L., Brodbelt J. S. (2009) Enhanced electron transfer dissociation through fixed charge derivatization of cysteines. Anal. Chem. 81, 7876–7884 [DOI] [PubMed] [Google Scholar]
- 50.Chalkley R. J., Medzihradszky K. F., Lynn A. J., Baker P. R., Burlingame A. L. (2010) Statistical analysis of Peptide electron transfer dissociation fragmentation mass spectrometry. Anal. Chem. 82, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao C., Turecek F. (2009) Host-guest hydrogen atom transfer induced by electron capture. J. Am. Soc. Mass Spectrom. 20, 639–651 [DOI] [PubMed] [Google Scholar]
- 52.Ledvina A. R., McAlister G. C., Gardner M. W., Smith S. I., Madsen J. A., Schwartz J. C., Stafford G. C., Jr., Syka J. E., Brodbelt J. S., Coon J. J. (2009) Infrared photoactivation reduces peptide folding and hydrogen-atom migration following ETD tandem mass spectrometry. Angew. Chem. Int. Ed. Engl. 48, 8526–8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.