Abstract
The mammalian spermatozoon has many cellular compartments, such as head and tail, permitting it to interact with the female reproductive tract and fertilize the egg. It acquires this fertilizing potential during transit through the epididymis, which secretes proteins that coat different sperm domains. Optimal levels of these proteins provide the spermatozoon with its ability to move to, bind to, fuse with, and penetrate the egg; otherwise male infertility results. As few human epididymal proteins have been characterized, this work was performed to generate a database of human epididymal sperm-located proteins involved in maturation. Two-dimensional gel electrophoresis of epididymal tissue and luminal fluid proteins, followed by identification using MALDI-TOF/MS or MALDI-TOF/TOF, revealed over a thousand spots in gels comprising 745 abundant nonstructural proteins, 408 in luminal fluids, of which 207 were present on spermatozoa. Antibodies raised to 619 recombinant or synthetic peptides, used in Western blots, histological sections, and washed sperm preparations to confirm antibody quality and protein expression, indicated their regional location in the epididymal epithelium and highly specific locations on washed functional spermatozoa. Sperm function tests suggested the role of some proteins in motility and protection against oxidative attack. A large database of these proteins, characterized by size, pI, chromosomal location, and function, was given a unified terminology reflecting their sperm domain location. These novel, secreted human epididymal proteins are potential targets for a posttesticular contraceptive acting to provide rapid, reversible, functional sterility in men and they are also biomarkers that could be used in noninvasive assessments of male fertility.
There are two major challenges for population and reproductive health. First, rapid global population expansion, which will see the world population reach 9 billion by 2045 (http://www.census.gov/ipc/www/idb/worldpopgraph.php). Second, infertility is suffered by around 180 million couples, comprising about 10%–15% worldwide and over 25% in some countries, of which 40%–50% is related to the male (http://www.who.int/reproductivehealth/publications/infertility/progress_63/en/index.html). Natural fertilization requires tightly coordinated and sequential sperm functions (1), from maturation in the epididymis (2) and survival in the female tract to fusion with the ovum (3). Fertility and infertility are both sides of the same reproductive coin so that understanding one can provide clues to the other. On the one hand, the widespread application to infertile couples of artificial reproductive technology, which bypasses natural fertilization mechanisms, has diverted attention from understanding the biological causes of the infertility for its cure, to that of overcoming the symptom of childlessness. On the other, controlling population growth by contraceptive means is currently only available to large populations of women. Male reproductive research lags far behind that of other disciplines and cannot meet the social needs for family planning, although hormonal contraception for men has been successful in China (4).
An alternative to suppression of sperm production for male contraception is to produce dysfunctional spermatozoa. Spermatozoa produced by the testis are immature, and since the epididymis provides a special microenvironment for their maturation and storage before ejaculation (5), it is a prime target for such attack. The concept of sperm maturation, first mooted by Young (6) as a time-dependent process, was challenged by Bedford (7) and Orgebin-Crist (8) who provided evidence for the role of the epididymis in this process. Subsequent studies characterized potentially important epididymal proteins. Rat epididymal secretory proteins B/C and D/E found in epididymal luminal fluids (9), bound to different sperm domains (10) and currently thought to regulate the process of capacitation (11), necessary for fertilization, have been cloned from a rat epididymal complementary DNA (cDNA)1 library (12). Other rat epididymal sperm-binding proteins, e.g. the antibacterial (13) Bin1B, are involved in sperm protection. Yanagimachi et al. (14) suggested that membranous apocrine secretions interact with hamster spermatozoa within the epididymal lumen and this has been demonstrated in the mouse (15). Human epididymal studies mirror the rodent work and confirm functional changes in epididymal spermatozoa (16) and specific proteins at the RNA and protein levels. The construction of human epididymal cDNA libraries from prostatic cancer patients led to the discovery of up to 12 human epididymal genes (17) that encode sperm-binding proteins with different functions (18). Dacheux et al. (19) performed two-dimensional gel analyses to characterize the human epididymal secretome and epididymosomes have been shown to transfer human epididymal proteins to spermatozoa (20).
Although the posttesticular approach offers rapid, effective, and reversible contraception, it requires interference with specific epididymal secretions (21, 22), but few such targetable secretions are known. The application of global cloning technology has defined the epididymal transcriptome of a fertile young man (23), which included transcripts found by gene chip arrays from organ donors (24, 25) and those in an epididymal cDNA library from elderly prostatic cancer patients (18). Far less is known of the epididymal proteome than transcriptome. Proteins in the epididymis of 15 species have been identified (26, 27), of which about 40 interact with spermatozoa, but few have defined functions that could be targeted for contraception. As molecular mechanisms can be manipulated to increase or reduce male fertility, knowing the proteins involved in and understanding the process of fertilization deserve more attention. To achieve this goal, this study aimed to produce a proteomic database and a high quality antibody library of human epididymal sperm proteins.
EXPERIMENTAL PROCEDURES
Experimental Design
The experimental design was to use normal adult epididymides from accident victims to release and separate luminal fluid from epididymal tissue minces. After removal of spermatozoa from both materials, proteins were extracted and analyzed for their identity and functional characteristics by two dimensional gel electrophoresis plus tandem mass spectrometry (MS/MS) technique and data mining. Antibodies were raised to these proteins and used to confirm the presence of the proteins in epididymal tissue and epididymal fluid by Western blotting, and to localize them on tissue sections by immunohistochemistry (IHC) and on initially motile ejaculated spermatozoa by immunofluorescence staining. Proteins that were found in tissue sections, detected in epididymal fluid, and present in spermatozoa were called epididymal secretory sperm-located proteins and the role of some representative proteins in sperm maturation was studied by sperm function tests using antibodies or recombinant proteins.
Human Organs and Ejaculated Semen for Immunolocalization
Epididymides from six fathers (27–32 years old), who had died in car accidents, were collected while artificial circulation maintained organs assigned for transplantation. Each epididymis was dissected into caput, corpus, and cauda regions, according to morphological features (Fig. S1). For total RNA extraction and IHC, one epididymis was frozen in liquid nitrogen. For protein extraction, the other epididymis was treated as follows. Normozoospermic semen (28) from 20 fathers (23–30 years) was obtained by masturbation after 7 days of sexual abstinence and was used for immunocytochemistry. Such a period of sexual abstinence increases sperm numbers without altering sperm motility, morphology, or vitality (29) and spermatozoa were obtained by swim-up to select the viable cells. All procedures were approved by the Ethics Committee of Yu-Huang-Ding Hospital.
Extraction of Epididymal Fluids and Tissue Proteins
Proteins were extracted (30) from all but the most proximal epididymal region, which comprises efferent ducts (31) (Fig. S1). Epididymal luminal fluid was obtained by decapsulating the epididymis and dividing it into caput, corpus and cauda regions. Luminal contents were released into phosphate-buffered saline containing protease inhibitors and EDTA by agitation at 4 °C for 20 min after coarse mincing and by applying gentle pressure to the tubule. Tubule fragments were removed by centrifugation at 130 × g for 5 min at 4 °C, and the supernatant further centrifuged at 5,700 × g to produce sperm-free “luminal fluid” fraction for protein extraction. Tubule fragments were washed twice without centrifugation, washed once and centrifuged at 6 × g and then at least once at 36 × g for 5 min until no spermatozoa were found after microscopic examination of the medium surrounding the last pellet. Sperm-free segments were ground to powder in liquid nitrogen, dissolved for 2 h in lysis solution (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm DTT) and centrifuged at 40,000 × g for 1 h at 4 °C. The supernatant and the luminal fluid were precipitated with 4 volumes of ice-cold acetone, stored for 1 h at −20 °C, thawed and centrifuged at 20,000 × g for 1 h at 4 °C. Precipitates were washed with 90% ice-cold acetone, dissolved in 5 ml lysis solution and stored at −80 °C. Sample protein concentrations were determined with the Bradford assay (32) (Bio-Rad).
Gel Electrophoresis, Mass Spectrometric Analyses, and Protein Identification
Two-dimensional gel electrophoresis was performed as previously described (33). Before the second dimensional electrophoresis, nonlinear pH 3–10 immobilized pH gradient strips (18 cm) used for one-dimensional isoelectric focusing were equilibrated in two steps: reduction with DTT and carboxymethylation with iodoacetamide. The equilibrated strips were run on 12% (w/v) SDS-PAGE at 25 mA per gel and stained with Coomassie Brilliant Blue R-350 (Amersham Biosciences). Gels were made in triplicate to confirm the spot patterns and were scanned with a Z320 scanner (Founder, Beijing, China). Gel images were processed with an Imagemaster (GE Healthcare). To detect low abundance proteins in the fluid fraction, one-dimensional Western blots were probed with each of the antibodies raised. Other proteins may not have been identified because of even lower abundance, limited by the sensitivity of the Western blotting (WB) method or imperfect MS data searching software to identify them.
Protein spots excised from two dimensional gels were destained with 25 mm NH4HCO3/50% (v/v) acrylonitrile and dried, followed by in-gel digestion with 0.01 μg trypsin in 0.01 ml 25 mm NH4HCO3 for 12 h at 37 °C. Digestion buffer was removed to a new 1.5-ml, clear microtube (Axygen Scientific, Union City, CA), 50 μl of 1% trifluoroacetic acid (v/v) in 50% (v/v) ACN was added to the gel plugs, which were sonicated for 30 min. This extract was removed and combined with the digestion buffer and freeze-dried (Labconco, Kansas City, MO) (34). Peptides were then resuspended in 15 μl of 0.5% (v/v) trifluoroacetic acid in Milli-Q water. Peptides were analyzed by a Voyager DE-STR biospectrometry work station (Applied Biosystems/MDS SCIEX, Foster City, CA) or a 4800 matrix-assisted laser desorption ionization (MALDI) time of flight (TOF)/TOF Analyzer (Applied Biosystems/MDS SCIEX) (35, 36). The MALDI-TOF mass spectrometer was operated in the delayed extraction/reflector mode with an acceleration voltage of 20 kV, a grid voltage setting of 72%, and a 120 ns delay. Positively charged ions were analyzed in reflection mode. External calibration was performed with the ProteomMass peptide and protein MALDI/MS calibration kit (Sigma). Each mass spectrum represents the sum of 150–200 laser shots collected from ≥30 different positions within each spot. Mass spectra were processed by Data explorer 4.0 (Applied Biosystems/MDS SCIEX). The parameters used were m/z range 800–4000, resolution >10,000, S/N threshold >10.0, and internal calibration by trypsin autodigestion peptides (trypsin_[108–115], MH+ 842.509; trypsin_[58–77], MH+ 2211.104). Masses frequently detected that arose from the matrix, trypsin, or known contaminants (e.g. keratins) were not analyzed. MALDI tandem mass spectrometry was performed on a TOF/TOF system (4800 Proteomics Analyzer, Applied Biosystems). The 4800 calibration mixtures (Applied Biosystems) were used to calibrate the spectrum to a mass tolerance within 150 ppm. For MS mode, peptide mass maps were acquired in positive reflection mode, and 800–4000 m/z mass range was used with 1000 laser shots per spectrum. A maximum of five precursors per spot with minimum signal/noise ratio of 50 were selected for MS/MS analysis. An energy of 2 KV was used for collision-induced dissociation (CID), and 2000 acquisitions were accumulated for each MS/MS spectrum. All of the automatic data analysis and database searching were fulfilled by the GPS ExplorerTM software (version 3.6, Applied Biosystems) running a mascot search algorithm (v2.1, Matrix Science, London, UK) for protein identification.
Data Mining and Bioinformatic Analysis
Database searches for MS was performed with Mascot (http://www.matrixscience.com/, MatrixScience) against the SWISS-PROT protein database (Swissprot Release 55.0; 356,194 sequences; 127,836,513 residues) for Homo sapiens. MS/MS was performed with GPS Explorer v.3.6. (Applied Biosystems/MDS SCIEX) programs, which incorporate the Mascot (v.2.1.) search algorithm (Matrix Science. Boston, MA) against the NCBInr database (NCBInr 20080210; 5,947,209 sequences; 2,045,123,248 residues) for Homo sapiens. The algorithm was set to use trypsin as the enzyme, allowing for one missed cleavage site and assuming carbamidomethyl as a fixed modification of cysteine and oxidized methionine as a variable modification. For protein mass fragment (PMF) data, peptide mass tolerance was set to ±0.3 Da. For MS/MS database searches, mass tolerance of precursor ions and fragment ions was set to 150 ppm and ±0.4 Da. Protein hits were considered identified if the Mascot score was greater than 60 and matched at least four peptides for peptide mass fingerprinting and 37 for MS/MS analysis (significance level, p < 0.05). If more than one protein was identified in a spot, the single protein member with the highest score (top rank) was chosen from the multiprotein family.
Proteins were distinguished functionally by a step-by-step classification and each protein was placed in only one category. The proteins were first scored according to their function reported in the literature and the Kyoto Encyclopedia of Genes and Genomes database (Release 52.0): proteins that could not be defined were sought in the PIR database (Release 15.9). Proteins were also scanned for protein domains by InterPro database (Release 23.0) and those with no annotation and supporting information were categorized as “Unclassified.”
Construction of the Antibody Library
To obtain a detailed mapping of potential epididymal secretory and sperm-located proteins, 628 antibodies against epididymal tissue pellet and fluid proteins were raised. For each protein, two to three peptide epitopes were designed to include isoforms identified in two-dimensional maps. In addition to several commercially available antibodies (151), most were prepared in-house by injecting male New Zealand White rabbits with recombinant proteins (18) or synthetic peptides (451), from which antisera or affinity-purified IgG were, respectively, prepared (titer, 1:10,000, Shandong Target Drug Research, Yantai, China). Because of the enormous effort involved, a listing of the successful peptide epitopes is withheld at present to safeguard the interest of the manufacturers. However, these antibodies and their peptide epitopes will be made available to interested scientists. In addition, six monoclonal antibodies were obtained by immunizing mice with recombinant proteins (see later discussion). Among the 230 nonstructural proteins of the 251 fluid proteins, 228 antibodies were usable. Of 494 tissue proteins, antibodies to 391 of 398 nonstructural proteins were usable. The quality of the antibodies against fluid and tissue proteins was checked by WB against their source. Of the 619 antibodies, 481 (78%) revealed only one band corresponding to the expected two-dimensional MW, 109 (18%) had more than one band, but with a major band corresponding to the targeted protein, and 29 (5%) had more than one band and no clear major bands, which may be isoforms.
Western Blotting and Reverse Transcription (RT)-PCR
Fluid and tissue samples (50 μg of each protein) separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes and blocked with 2% (w/v) skimmed milk for 1 h. Thereafter they were incubated for 1 h with the primary antibody at room temperature (RT) with gentle agitation. After several washes with 0.5% (v/v) Tween-20 in Tris buffered saline, membranes were incubated for 1 h at RT with HRP-conjugated anti-IgG at a final dilution of 1:700 in blocking solution. Immunoreactive complexes were visualized using a diaminobenzidene kit (ZhongShan Biotechnology, Beijing, China).
Total RNA from human tissues was extracted with Trizol (Tianwei, Beijing, China) following the manufacturer's recommendations. cDNAs were synthesized according to instructions provided with the avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI). Forward and reverse primers for the genes of epididymal proteins were designed from Primer Premier 5.0 Software (PREMIER Biosoft International, Palo Alto, CA) on the basis of sequences published in GenBank and are listed in Table S1. PCR was performed with a PE9700 (Applied Biosystems) and PCR products were separated on a 1% (w/v) agarose gel, visualized by staining with ethidium bromide and detected by a KODAK EDAS120 system (Carestream Health, New Haven, CT). To check for epididymal specificity, all genes were examined by RT-PCR in other organs from accident victims (heart, liver, spleen, lung, and kidney).
Immunohistochemistry and Immunocytochemistry
Tissues were frozen in liquid nitrogen after their isolation from the donor and then embedded in Tissue OCT-Freeze Medium (Leica, Nussloch, Germany). Air-dried cryosections (6 μm) were placed on gelatin-coated slides, air-dried, fixed with ice-cold acetone for 10 min and permeabilized for 10 min with Tween-20 in Tris buffered saline. Sections were incubated with 1% (v/v) hydrogen peroxide (H2O2) and washed in Tween-20 in Tris buffered saline. Sections were blocked for 1 h with blocking solution (3% (w/v) bovine serum albumin (BSA) in Tris buffered saline) at RT and subsequently incubated at 37 °C for 2 h with primary antibody (diluted 1:50 in blocking solution). After several washes with Tris buffered saline, sections were incubated for 60 min at RT with horse radish peroxidase-conjugated anti-IgG (ZhongShan Biotechnology, Beijing, China) at a final dilution of 1:200 in blocking solution. Peroxidase activity was revealed by a diaminobenzidine kit (ZhongShan Biotechnology). Sections were counterstained with hematoxylin and dehydrated, mounted for bright-field microscopy with DM LB2 (Leica, Nussloch, Germany). Pre-immune serum (rabbit, mouse, and goat) was used as negative control and anti-β-actin (Santa Cruz Biotechnology, Santa-Cruz, CA), anti-HEL-S-154w (previously anti-HEL-75 (37) sera were used as positive controls.
Thirty min after liquefaction, human ejaculated spermatozoa were swum-up in M199 and the motile cells washed once in phosphate buffered saline (PBS). The sperm pellet was placed on 1% (w/v) gelatin-coated slides, air-dried, and fixed with ice-cold methanol for 10 min. Slides were blocked for 1 h at RT with 3% (w/v) BSA in PBS and incubated at 37 °C for 30 min with antibodies (diluted 1:50 in PBS containing 3% (w/v) BSA). After three washes with PBS, the corresponding secondary antibody was applied (FITC-labeled IgG, 1:200 in PBS containing 3% (w/v) BSA). Samples were subsequently washed in PBS and deionized water. Propidium iodide (0.01 mg/ml; Invitrogen, Carlsbad, CA) counterstaining visualized the nuclei. After the staining, all sections were mounted in 80% (v/v) glycerol and examined with a confocal laser scanning microscope (LSM-510 META; Carl Zeiss, Jena, Germany). Preimmune serum was used as a negative control and anti-RNase9 serum (38) as a positive control.
Effect of Antibodies against HEL-S-124m and HEL-S-89n on Sperm Motility
After liquefaction at 37 °C for 30 min, spermatozoa were allowed to swim up into M199. After centrifugation at 500 × g for 5 min the sperm pellet was washed twice with Biggers-Whitten-Whittingham buffer containing 35 mg/ml BSA and 25 mm bicarbonate and the sperm concentration adjusted to 27 × 106/ml. Sperm suspensions were incubated for 3 h to allow capacitation, when samples were divided into two aliquots. Irrelevant IgG (0.02 mg/ml; Sigma) was added to control tubes and affinity-purified antibody IgG (also 0.02 mg/ml) was added to the test samples. Both samples were incubated for 1 h at 37 °C in 5% (v/v) CO2. Aliquots were subsequently placed in 20-μm deep chambers (2X-CEL, Hamilton Thorne Research, Beverly, MA) and motility was measured at 37 °C by computer-aided sperm analyzer (HTM-IVOS, Version 12.3, Hamilton Thorne Research).
Effect of HEL-S-128m on Peroxidative Stress of Spermatozoa
Spermatozoa collected by swim-up as described previously. The sperm pellet was resuspended in Tris buffered saline (pH 7.2) and cell concentration was adjusted to 3 × 106/ml. In the experimental group, various concentrations of HEL-S-128m (0.0625, 0.125, 0.25, 0.5, and 1.0 mg/ml) were added together with H2O2 (200 and 800 μm), whereas in the control group only H2O2 was added. Samples were then incubated for 30 min at 37 °C in 5% CO2. Motility was then analyzed as described earlier.
RESULTS
Number and nature of proteins in epididymal fluid and tissue
The quality of the two-dimensional gels (Fig. 1a, b) permitted detection of more than 1720 protein spots, of which 512 were in the fluids and 1208 in tissue pellet. A total of 1508 protein species could be identified by MALDI-TOF/MS and MALDI-TOF/TOF (Table S2). A total of 251 unique proteins from 512 protein spots were found in epididymal luminal fluid and 622 unique proteins from 1208 protein spots in the epididymal tissue pellet. There were 128 proteins common to both epididymal fluid and tissue, suggesting that 51% of the abundant luminal fluid proteins are also present as abundant proteins in the tissue, whereas 21% of abundant proteins in the tissue were detected as abundant proteins in the fluid. Thus, the human epididymal proteome presented here comprises 745 unique proteins in fluid and tissue (251 + 622 − 128), of which 117 were structural proteins (Table S3), which were not further studied.
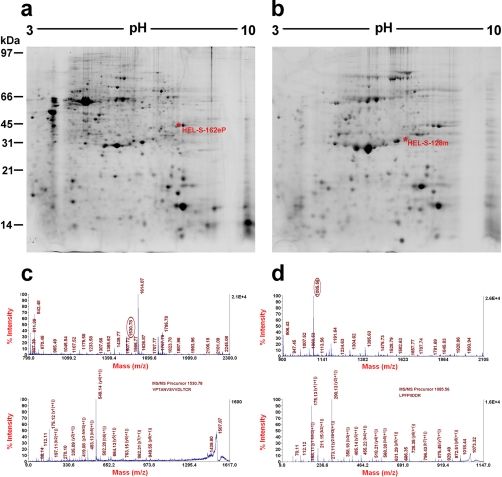
Fig. 1.
Separation and identification of human epididymal proteins by 2D-PAGE and MALDI-MS Spectra. Reference maps of (A) the epididymal tissue pellet; (B) the epididymal fluid; example spectra of (C) HEL-S-162eP, (D) HEL-S-128m. The MS map (C, upper panel) and MS/MS map (C, lower panel) marked with b ions and y ions for glyceraldehyde-3-phosphate dehydrogenase identification. The sequence of precursor at m/z 1530.78 was analyzed by MS/MS to be VPTANVSVVDLTCR and the protein identified as glyceraldehyde-3-phosphate dehydrogenase. The MS map (D, upper panel) and MS/MS map (D, lower panel) marked with b ions and y ions for peroxiredoxin 6 identification. The sequence of precursor at m/z 1530.78 was analyzed by MS/MS to be LPFPIIDDR and identified as peroxiredoxin 6.
In total, 628 antibodies (230 against luminal fluid proteins and 398 against tissue pellet proteins) were raised against nonstructural proteins and all were confirmed by WB against their source (luminal fluids or tissue pellet proteins) (Fig. S2), and 619 antibodies (98%) were positive. These were used to locate the proteins in tissue and luminal fluid preparations (Fig. S3). In addition to the 228 abundant two-dimensional fluid proteins with positive WB results, an additional 180 low abundance proteins identified by antibodies raised against tissue pellet two-dimensional proteins were detected in luminal fluids by one-dimensional WB, revealing that the fluid contained over 400 different proteins.
Expression of gene transcripts and translation products
Immunohistochemical analysis revealed that most of the proteins identified were associated with epithelial cells lining the epididymal duct, rather than with peritubular or interstitial cells (Fig. 2). For each protein, immunological expression in different epididymal segments varied and many were on the microvilli (Fig. S4), although not all of these were detected in fluids by Western blotting. Analysis of each protein's transcript by RT-PCR confirmed the regional differences in epididymal expression and also demonstrated expression of some of these in other organs (Table S4). There were 40 of the 207 sperm-located proteins (36 also in epididymal fluid) that had negative testicular RT-PCR results (see highlights in Table S5).
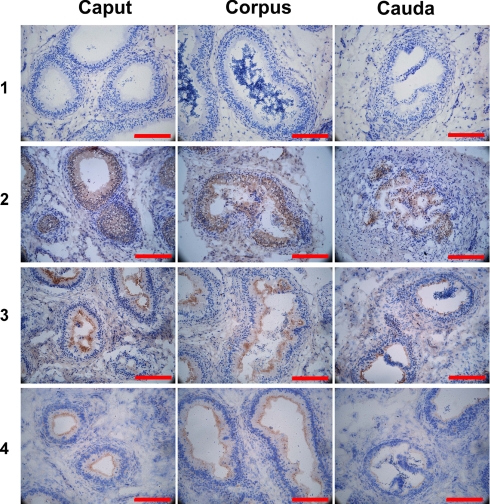
Fig. 2.
Immunohistochemical location of sperm-located and nonlocated proteins in tissue sections. Representative examples of proteins located in different epididymal regions (left panels, Caput; middle panels, Corpus; right panels, Cauda) in different tissue compartments. 1, negative control, preimmune rabbit serum; 2, secreted, sperm-bound HEL-S-97n, in the epididymal epithelium of all three regions; 3, secreted, HEL-S-153, in the epididymal epithelium and on microvilli of all three regions; 4, nonsecreted HEL-T-60, in the epididymal epithelium and on microvilli of all three regions. Each bar represents 200 μm.
There was general agreement between RT-PCR and IHC results but there were some differences. For example, of the 15 proteins localized in the three epididymal regions by IHC listed in Table I, RT-PCR failed to detect transcripts in only two cases: for proteins HEL-S-55e in the corpus and HEL-S-123m in the caput. In the former case the protein may have been synthesized in the caput, upstream of the sampled site, but taken up into the tissue at the corpus. In the latter case, the IHC staining was very weak.
Table I. Proteomic data of examples of epididymal secretory sperm-located proteins representing each location pattern as displayed in Fig. 3.
Each pattern number designates a protein location on spermatozoa. Data of all such 174 secretory sperm-located proteins plus 33 nonsecretory sperm-located proteins and all 619 epididymal proteins are given in supplemental Table 5. Each protein was also localized by IHC (see supplemental Fig. 4) in the three epididymal regions of caput, corpus, and cauda and the expression of their transcripts was confirmed by RT-PCR (see supplemental Table 4). ** Percentages of antibody-positive cells: +++, >75%; ++, 51%–75%; +, 25%–50%; ±, <25%. *** absence of mRNA, but presence of protein, in the cauda may reflect uptake of protein synthesized and secreted upstream rather than de novo synthesis. ê, in-house antibodies.
| Protein location |
Total protein number | Example HEL-S-nx | Gene symbol | Protein name | MW (kDa) | pI | Chrom. No. | Antibody host |
Protein expression** |
Gene expression |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern no. | Sperm Domain | Goat | Rabbit | Mouse | Caput | Corpus | Cauda | Caput | Corpus | Cauda | |||||||
| 1 | Acrosome | 40 | 25a | STAT5A | Signal transducer and activator of transcription 5A | 91 | 5.98 | 17 | √# | + | ++ | ++ | ++ | + | + | ||
| 2 | Equatorial | 15 | 55e | GLYAT | Glycine N-acyltransferase) | 34 | 8.38 | 11 | √# | ++ | ++ | + | + | - | + | ||
| 3 | Post-acrosomal | 28 | 69p | PPIA | Peptidyl-prolyl cis-trans isomerase A | 18 | 7.68 | 7 | √ | + | ± | ± | +++ | + | + | ||
| 4 | Neck | 30 | 97n | PRDX4 | Peroxiredoxin-4 | 31 | 5.86 | X | √ | ++ | +++ | ++ | ++ | + | +++ | ||
| 5 | Midpiece | 10 | 123m | ATP5A1 | ATP synthase subunit α, mitochondrial precursor | 44 | 9.42 | 18 | √ | + | ++ | +++ | - | ++ | -*** | ||
| 6 | Princpal piece | 20 | 133p | LDHA | l-lactate dehydrogenase A chain | 37 | 8.46 | 11 | √ | ++ | +++ | +++ | + | +++ | +++ | ||
| 7 | End-piece | 3 | 150e | COL6A1 | Collagen α-1 chain precursor | 108 | 5.26 | 21 | √# | + | ++ | ± | ++ | +++ | + | ||
| 8 | Whole sperm | 2 | 154w | DEFB132 | β-defensin 132 precursor | 8 | 9.62 | 20 | √# | ++ | +++ | + | ++ | ++ | ++ | ||
| 9 | Acrosome + Neck | 3 | 157an | FBN1 | Fibrillin-1 precursor | 312 | 4.81 | 15 | √# | ± | + | + | +++ | +++ | +++ | ||
| 10 | Acrosome + Midpiece | 2 | 158am | CTSB | Cathepsin B precursor | 23 | 5.19 | 8 | √# | ++ | ++ | +++ | + | ++ | +++ | ||
| 11 | Acrosome + Principal piece | 2 | 160ap | HSPA9 | HSPA9 protein | 74 | 6.04 | 5 | √# | ++ | ++ | ++ | ++ | ++ | +++ | ||
| 12 | Equatorial + Principal piece | 1 | 162ep | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 36 | 8.26 | 12 | √ | ++ | +++ | ++ | +++ | ++ | ++ | ||
| 13 | Postacrosomal + Annulus | 1 | 163pA | A1BG | α-1B-glycoprotein precursor | 54 | 5.58 | 19 | √ | + | + | + | ++ | +++ | ++ | ||
| 14 | Neck + Annulus | 1 | 164nA | GANAB | Neutral α-glucosidase AB precursor | 107 | 5.74 | 11 | √ | + | ++ | ++ | ++ | +++ | +++ | ||
| 15 | Midpiece + Prinicipal piece | 16 | 181mP | SAMD8 | Sphingomyelin synthase-related protein 1 | 48 | 8.35 | 10 | √ | ++ | +++ | + | + | +++ | ++ | ||
Protein Location in Mature Spermatozoa
The antibody library permitted each epididymal protein to be located on swim-up spermatozoa where 15 combinations of antibody location were found (Fig. 3). In total 207 sperm-located proteins were identified (Fig. 4).
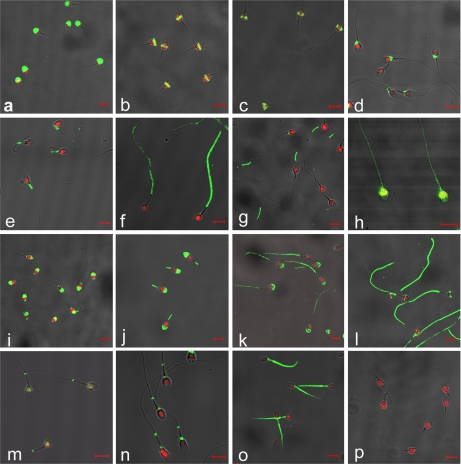
Fig. 3.
Immunocytochemistry of washed ejaculated human spermatozoa incubated with antibodies against epididymal secretory proteins. Merged micrographs of fluorescence staining. Proteins selected for association with specific sperm domains: a, acrosome' HEL-S-25a65307; b, equatorial region'HEL-S-55e65307; c, postequatorial region'HEL-S-69p65307; d, neck'HEL-S-97n65307; e, midpiece'HEL-S-123m65307; f, principal piece'HEL-S-133P65307; g, end piece'HEL-S-150E65307; h, whole sperm'HEL-S-154w (previously HEL-75)65307; i, acrosome and neck'HEL-S-157an65307; j, acrosome and midpiece'HEL-S-158am65307; k, acrosome and principal piece'HEL-S-160aP65307; l, equatorial and principal piece'HEL-S-162eP65307; m, postequatorial and annulus'HEL-S-163pA65307; n, neck and annulus'HEL-S-164nA65307; o, midpiece and principal piece'HEL-S-181mP; p, pre-immune serum as negative control. Red staining (PI) indicates the nuclei, green staining (FITC) indicates the location of epididymal secretory proteins on spermatozoa. Each bar represents 5 μm.
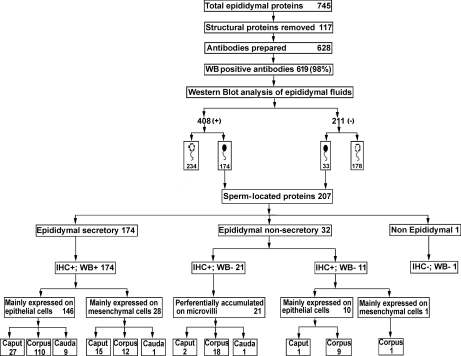
Fig. 4.
Flow chart of identification and categorization of the 207 sperm-related proteins from 745 initially identified tissue and fluid proteins. The number of proteins examined was reduced by ignoring 117 structural proteins (unlikely to be secreted) and antibodies were raised to the remaining 628 proteins with 98% success. With the 619 usable antibodies, Western blots revealed that 408 proteins were present in epididymal fluid and 211 not. Two hundred and seven proteins were sperm-located (174 proteins found in epididymal fluid, 33 not) and the remaining proteins (412) were not associated with spermatozoa, whether found in fluid (234) or not (178). Filled sperm head, sperm-located proteins; clear sperm head, non sperm-located proteins; IHC: immunohistochemistry; WB: western blots of epididymal luminal fluid; +65306 positive staining 65307 −65306 negative staining.
Nomenclature of epididymal proteins
On the basis of WB and immunostaining results, proteins were classified as tissue proteins, secretory proteins, and sperm-located proteins. Nonsecretory tissue proteins were defined as those present in the tissue pellet but not in epididymal fluid; secretory proteins were defined as those present in epididymal fluid; and sperm-located proteins were those whose antibodies detected signals on washed ejaculated spermatozoa. The terminology for nonsecretory tissue proteins was human epididymal protein-li tissue (HEL-T-n), where n identifies the specific protein, and for secretory proteins human epididymal protein-li secretory (HEL-S-n). Depending on their location on spermatozoa, sperm-located proteins were further subclassified into 15 groups with names ending in a representative letter (x) relating to the protein's location on the spermatozoon. Such sperm-located proteins were designated HEL-S-nx for secretory and HEL-T-nx for nonsecretory sperm-located proteins where x could be (a) sperm acrosome, (e) equatorial region, (p) postacrosomal, (n) neck, (m) midpiece, (A) annulus, (P) principal-piece, (E) end-piece or their combinations. Table I lists examples of 15 proteins that are located at 15 sperm domains. Each sperm-located protein received an individual nomenclature that was registered with GenBank (http://www.ncbi.nlm.nih.gov/Genbank).
Details of all the sperm-located, secreted and tissue proteins are presented in Table S5, which lists a total 619 epididymal proteins whose antibodies were used to detect the proteins by Western blots in epididymal luminal fluid, by immunohistochemistry in cells in various regions of the epididymis, and by immunocytochemistry on different domains of ejaculated spermatozoa. Of these proteins there were 234 epididymal, secretory nonsperm-located proteins (HEL-S-n); 174 epididymal, secretory sperm-located proteins (HEL-S-nx); 33 nonsecreted tissue sperm-located proteins (HEL-T-nx), and 178 nonsecreted nonsperm-located proteins (HEL-T-n) of which 16 were found on the microvilli of the epithelium. Some of the sperm-located proteins have been identified by other workers as being on the sperm surface (Table S6).
Molecular Weight, Isoelectric Point and Chromosomal Location of Epididymal Proteins
The isoelectric points of the human epididymal proteins ranged from 4.5 to 9 (Fig. S6) and the molecular weights of the identified human epididymal proteins were under 100 kDa with most between 20 and 50 kDa (Fig. S7). The chromosomal location of the epididymal genes transcribing the secretory proteins and sperm-located proteins were localized on the autosomes or on the X-, but not on the Y-, sex chromosome (Fig. S8).
Classification of Biological Function
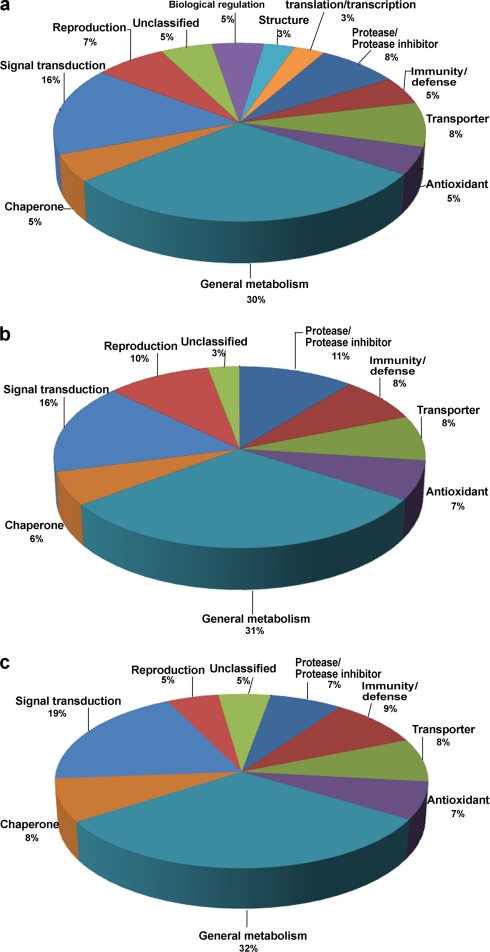
The proteins distinguishable as spots on two-dimensional gels (Fig. 1A, B) and identified by mass spectrometry were placed into broad functional categories on the basis of the KEGG and PubMed databases. The majority (30%) of all proteins were related to metabolism (Fig. 5) whereas other protein functions were related to different sperm locations. Table S7 shows the percentage of 207 proteins found on different sperm domains that are associated with different putative functions.
Fig. 5.
Pie diagrams of the proportion of epididymal proteins categorized by function. Functional classification of (A) all 745 epididymal proteins, (B) 408 epididymal secretory proteins, (C) 207 epididymal sperm-located proteins.
Effect of Antibodies against HEL-S-124m and HEL-S-89n on Sperm Motility
The antibody to HEL-S-89n (78-kDa glucose-regulated protein precursor) decreased the percentage of progressive motility and swimming velocities of capacitated spermatozoa from control values obtained in the presence of an equal concentration of irrelevant IgG. The antibody to HEL-S-124m (stress-70 protein, mitochondrial precursor) did not alter swimming velocities significantly but did change the swimming pattern amplitude of lateral head displacement (ALH) from that of the control (Table II).
Table II. Effect of antibodies against selected secreted human epididymal proteins located on the sperm tail on the values of kinematic parameters. Antibodies against HEL-S-89n (78 kDa glucose-regulated protein) located at the sperm neck and HEL-S-124m (Stress-70 protein, mitochondrial precursor) located in the midpiece, modify the kinematic parameters of capacitated spermatozoa compared with IgG-treated controls. VAP, averaged path velocity; VSL, rectilinear velocity; VCL, curvilinear velocity; ALH, amplitude of lateral head displacement; BCF, beat cross frequency; STR = VAP/VCL; LIN = VSC/VCL. *, within each row, significantly different from control. For each antibody, between 476 and 816 motile spermatozoa in total were assessed in either the control or experimental groups in five experiments.
| Parameter | Control | HEL-S-89n | P | Control | HEL-S-124m | P |
|---|---|---|---|---|---|---|
| Total motility (%) | 70.0 ± 12.8 | 60.0 ± 18.1 | 0.74 | 61.3 ± 4.0 | 39.7 ± 10.2 | 0.06 |
| Progressive motility (%) | 39.5 ± 14.6 | 31.8 ± 11.6* | 0.02 | 28.3 ± 2.5 | 21.7 ± 8.0 | 0.27 |
| VAP (μm/s) | 67.8 ± 10.2 | 61.5 ± 10.5* | 0.01 | 64.3 ± 9.1 | 49.0 ± 5.6 | 0.11 |
| VSL (μm/s) | 52.5 ± 10.2 | 47.3 ± 8.4* | 0.02 | 47.0 ± 9.1 | 38.7 ± 5.7 | 0.21 |
| VCL (μm/s) | 132.0 ± 12.6 | 122.3 ± 3.5 | 0.07 | 128.3 ± 12.1 | 96.0 ± 13.2 | 0.12 |
| ALH (μm) | 6.3 ± 0.4 | 6.4 ± 0.4 | 0.71 | 6.3 ± 0.6 | 5.0 ± 0.7* | 0.01 |
| BCF (Hz) | 25.8 ± 1.3 | 26.0 ± 2.2 | 0.76 | 26.3 ± 0.6 | 24.0 ± 2.0 | 0.12 |
| STR (%) | 77.8 ± 3.4 | 77.3 ± 3.1 | 0.64 | 74.7 ± 2.9 | 78.3 ± 3.2 | 0.09 |
| LIN (%) | 41.8 ± 4.5 | 41.5 ± 3.7 | 0.64 | 39.0 ± 1.7 | 41.7 ± 2.5 | 0.09 |
Effect of HEL-S-128m on Peroxidative Stress of Spermatozoa
Incubation of capacitated spermatozoa with H2O2 alone (200 μm) compromised almost all motile activity but in the presence of 0.5 mg/ml HEL-S-128m (peroxiredoxin-6) there were 54% fast swimming spermatozoa. A higher H2O2 concentration (800 μm) alone totally abolished sperm motility but 1 mg/ml HEL-S-128m rescued motility to 80% fast swimming spermatozoa (for each trial n = 3, p < 0.05, one way ANOVA). The data in Fig. S9 reveal the dose-dependent increase in percentage of fast progressive spermatozoa and decrease in percentage of immotile spermatozoa with increasing concentrations of HEL-S-128m.
DISCUSSION
Now that the human genome project has ended, several human proteomic projects have begun, such as the multi-center collaborations on the liver proteome (39, 40, 41). The systematic use of such engineering proteomic techniques will permit a database of human epididymal proteins to be constructed for use as markers for diagnosis and therapy for infertility.
The complex nature of sequential addition, removal, and modification of secreted testicular and epididymal proteins on the surface of testicular spermatozoa during their epididymal transit has until now been sketchy. Human epididymal spermatozoa are not readily available (most material comes from old men with prostatic carcinoma; organs from young cancer patients, or organ donations are rare; and for none is the current fertility status known) and biopsies cannot be taken without compromising fertility. Here good quality epididymal tissue was used, from fathers who had died in accidents, and a large novel database of human epididymal proteins was created from systematic work-up of over a thousand proteins, the use of over 600 antibodies and confirmation by transcript detection. The proteomes of mature epididymal spermatozoa from mice and rats, but not from men, have been published (42, 43). The proteome of human ejaculated spermatozoa has been studied by the use of sensitive silver-staining, antibodies found in serum from infertile patients, one-dimensional gels and LC-MS/MS (one fertile man) (44), and two-dimensional gels and MALDI-TOF plus electrospray ionization-ion trap (11 normozoospermic men) (45). There is no proteomic study of the epididymal contribution to sperm-maturation antigens (see reviews 46, 47) and our interest was focused on proteins of epididymal origin because they are necessary for sperm maturation and are potential posttesticular contraceptive targets. The specific functions and locations of the proteins on functionally specific sperm domains presented here will open a new area of functional research.
Of the 619 antibodies raised to human epididymal proteins, the interaction of 207 with washed ejaculated spermatozoa indicates that the size of the human epididymal sperm-located protein family is much larger than previously envisaged. Before this publication, 24 such human epididymal proteins had been identified, registered, published, and named by international centers. They were found in different laboratories, by methods different from the technology used here, which also identified them. Most of the proteins reported here were considered epididymal secretory proteins from their presence in epididymal fluid but the transcripts for some were expressed by other organs. However, as some epididymal protein transcripts were found in the testis, it is possible that such proteins in the epididymal fluid originate from the testis alone or also from the epididymis. The location on spermatozoa of some proteins observed here differed from those reported; this may reflect different antibodies, methodology, or sperm populations, and requires further investigation.
Most of the proteins reported here were considered epididymal secretory proteins from their presence in epididymal fluid but they are not epididymal-specific since transcripts were expressed by other organs. The presence of proteins in epididymal fluid is not definitive proof of an epididymal origin, since they could also have arisen in the testis or have dual origins in both the testis and epididymis. Nevertheless, the expression of 36 of the HEL-S-nx proteins at the testicular mRNA level was nondetectable, substantiating their epididymal origin. On the other hand, another 33 epididymal sperm-located proteins could not be confirmed as secreted because they were nondetectable in the luminal fluid; these may be present below the sensitivity of the Western blotting method. Whereas uptake of proteins into the sperm cell cytoplasm has not yet been demonstrated, our immunocytochemical method was unable to differentiate proteins on the membrane from those in the cytoplasm so the proteins located in spermatozoa are termed “sperm-located proteins.“ To prove that such proteins are of posttesticular origin, examination of whole testicular spermatozoa is necessary. Whereas isolation of sufficient free testicular spermatozoa is difficult, we are currently attempting to obtain human epididymal spermatocoeles, which contain free testicular spermatozoa.
The total number of proteins in a spermatozoon is estimated to be around 2000–2500 (48). The present work identified 174 proteins, expressed by epididymal tissue and present in luminal fluid, that were sperm-located and there is a fair chance that these proteins are of epididymal origin. The peculiar nature of apocrine secretion in the epididymis, with shedding of apical blebs of cytoplasm into the lumen that liberate luminal vesicular epididymosomes (49) explains how proteins lacking a signal sequence can be secreted into the epididymal lumen. The specific transfer of glycosylphosphatidyl inositol anchored proteins in lipid rafts in the vesicles to the sperm membrane has been demonstrated in bulls (49). The proteome of human epididymosomes, consisting of 146 proteins, has been published (50) and there were overlaps with 26 (11%) of the 234 luminal proteins (HEL-S-n) and with 23 (11%) of the 207 sperm-located proteins (HEL-S-nx) reported in this study.
In rodents the differences between caput and cauda epididymidal sperm proteins reveal those important during sperm maturation (51). Many of the epididymal secretory proteins found on the washed ejaculated spermatozoa here are also found in the proteomes of human and rodent spermatozoa to represent proteins essential for sperm development or function. For instance, of the list of 16 chaperone proteins (48) (which assist the noncovalent folding/unfolding and the assembly/disassembly of macromolecules, and prevent aggregation of newly synthesized or modified polypeptides) five were identified in this study. These include the two heat shock 70-kDa proteins, HSP90-α, clusterin, and protein disulfide-isomerase A3 precursor. By differential in-gel electrophoresis analysis in rats, HSP70 is one of three sperm proteins reported to increase after passage from caput to cauda epididymidis (51), indicative of epididymal secretion and uptake by spermatozoa. Indeed, the homologue gene Hspa4 in the rat is highly expressed in the proximal segments of the epididymis (52).
The present bioinformatic analysis located all the epididymal proteins to the autosomes and the X, but not Y, chromosome, in agreement with previous results from the human epididymal transcriptome (23, 53). Functional analysis shows that epididymal proteins control the luminal microenvironment and sperm maturation (54). These sperm-coating metabolic proteins may be related to metabolism required during maturation or at fertilization (55), especially if related to membrane enzyme or ion transport activity. The ability of antibodies against proteins located at the neck and midpiece of the flagellum to modify sperm motility, indicates not only that the proteins are on the sperm surface, but that they may be instrumental in regulating some aspects of flagellar function. Although the ability of PRDX6 to prevent H2O2-induced damage is explained by its known function, this is not apparent from their proteomic identification as glucose-regulated protein precursor-binding protein grp78 and the mitochondrial precursor of stress-70 protein, they could nevertheless serve as targets for contraception.
More than 20% of the proteins were involved in immune defense, antioxidation and molecular chaperoning, including the protein HEL-75 (renamed HEL-S-154w here) already reported (37). The surface proteins may well participate in immunological protection of epididymal spermatozoa in the male and female tracts. The ability of the protein HEL-S-128m to prevent the hydrogen peroxide-induced reduction in motility, confirms its proteomic identification as periredoxin-6, a hydroperoxidase, and suggests a function as an antioxidant within the male tract.
Of the epididymal secretions found on spermatozoa, about 19% were signal transduction proteins that may be important for regulating sperm functions during the process of epididymal sperm transit and fertilization (55). The functional classification of proteins located on different sperm domains revealed that proteins binding to the sperm neck had mainly immunological functions, whereas those binding to the sperm acrosome, equatorial region, and tail had signal transduction functions. The former may be related to the survival of spermatozoa as foreign cells in the female tract and the latter to the sequential events that the sperm head and tail undergo at fertilization, including hyperactivated motility, zona-binding, acrosomal exocytosis, zona penetration, oolemma-binding, and fusion (56).
This work has greatly expanded the human epididymal proteome and classified a large group of sperm maturation-related proteins into a family by a systematic nomenclature that reflects the position of each protein on spermatozoa. The epididymal secretory sperm-located proteins are important proteins in sperm maturation and the finding that proteins of different function are preferentially located at different sperm domains will open a new area of functional research. Several reports indicate that corresponding sperm functions are compromised by antibodies against proteins from the sperm head and tail (57, 58, 59, 60).
In view of the diversity of sperm functions, this database of well-characterized sperm proteins should serve as a source of reference for future research and clinical application. The roles of the new epididymal proteins have to be substantiated before they can be used to develop diagnostic tests, realize therapeutic possibilities, and become contraceptives for men. From a public health perspective, this work will permit men to play an important role in family planning to the improvement of life quality.
Supplementary Material
Acknowledgments
We thank Guo Lihai PhD (Shanghai Asia Pacific Application Support Center, Applied Biosystems, China) for help in identifying two-dimensional gel spots with a 4800 MALDI TOF/TOF Analyzer; Ji Zhiliang PhD (Xiamen University, bioinformatics-aided drug discovery group, China) for advice on bioinformatic analysis. Dr Trevor G. Cooper and Dr Ching-Hei Yeung (Centre of Reproductive Medicine and Andrology, University of Muenster, Germany) for critical review and editing of the manuscript. Pro Shi QiXian (Zhejiang Academy of Medical Sciences, Hangzhou, China) for review and advice of the manuscript. We declare no competing interests.
Footnotes
* This work was supported by grants from China National 973 Programme Project (2009CB521704).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- cDNA
- complementary DNA
- WB
- Western blotting
- IHC
- Immunohistochemistry
- RT
- room temperature
- MS/MS
- tandem mass spectrometry
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MALDI
- matrix-assisted laser desorption ionization
- TOF
- time of flight
- RT-PCR
- reverse transcription-PCR
- BSA
- bovine serum albumin
- PBS
- phosphate buffered saline.
REFERENCES
- 1.Yanagimachi R.Mammalian fertilization (1994). In The Physiology of Reproduction. 2nd Ed.Knobil E., Neill J. D., (eds.) Raven, New York, p. 189 [Google Scholar]
- 2.Cooper T. G., Yeung C. H. (2006). Sperm maturation in the human epididymis. In The Sperm Cell: Production, Maturation, Fertilization, Regeneration. De Jonge C, Barratt C. L. R. (eds.) CUP, Cambridge, UK, pp. 72 [Google Scholar]
- 3.Suarez S. S., Pacey A. A. (2006) Sperm transport in the female reproductive tract. Hum Reprod Update 12, 23–37 [DOI] [PubMed] [Google Scholar]
- 4.Gu Y., Liang X., Wu W., Liu M., Song S., Cheng L., Bo L., Xiong C., Wang X., Liu X., Peng L., Yao K. (2009) Multicenter contraceptive efficacy trial of injectable testosterone undecanoate in Chinese men. J. Clin. Endocrinol. Metab. 94, 1910–1915 [DOI] [PubMed] [Google Scholar]
- 5.Turner T. T. (1995) On the epididymis and its role in the development of the fertile ejaculate. J. Androl. 16, 292–298 [PubMed] [Google Scholar]
- 6.Young W. C. (1931) A study of the function of the epididymis. III. Functional changes undergone by spermatozoa during their transit through the epididymis and vas deferens in the guinea pig. J. Exp. Biol. 8, 109–210 [Google Scholar]
- 7.Bedford J. M. (1967) Effects of duct ligation on the fertilizing ability of spermatozoa from different regions of the rabbit epididymis. J. Exp. Zool. 166, 271–281 [DOI] [PubMed] [Google Scholar]
- 8.Orgebin-Crist M. C. (1967). Fertility in does inseminated with epididymal spermatozoa. J. Reprod. Fertil. 14, 346–347 [DOI] [PubMed] [Google Scholar]
- 9.Brooks D. E., Higgins S. J. (1980) Characterization and androgen-dependence of proteins associated with luminal fluid and spermatozoa in the rat epididymis. J. Reprod. Fertil. 59, 363–375 [DOI] [PubMed] [Google Scholar]
- 10.Brooks D. E., Tiver K. (1983) Localization of epididymal secretory proteins on rat spermatozoa. J. Reprod. Fertil. 69, 651–657 [DOI] [PubMed] [Google Scholar]
- 11.Roberts K. P., Johnston D. S., Nolan M. A., Wooters J. L., Waxmonsky N. C., Piehl L. B., Ensrud-Bowlin K. M., Hamilton D. W. (2007) Structure and function of epididymal protein cysteine-rich secretory protein-1. Asian J. Androl. 9, 508–514 [DOI] [PubMed] [Google Scholar]
- 12.Brooks D. E., Means A. R., Wright E. J., Singh S. P., Tiver K. K. (1986) Molecular cloning of the cDNA for androgen-dependent sperm-coating glycoproteins secreted by the rat epididymis. Eur. J. Biochem. 161, 13–18 [DOI] [PubMed] [Google Scholar]
- 13.Li P., Chan H. C., He B., So S. C., Chung Y. W., Shang Q., Zhang Y. D., Zhang Y. L. (2001) An antimicrobial peptide gene found in the male reproductive system of rats. Science 291, 1783–1785 [DOI] [PubMed] [Google Scholar]
- 14.Yanagimachi R., Kamiguchi Y., Mikamo K., Suzuki F., Yanagimachi H. (1985) Maturation of spermatozoa in the epididymis of the Chinese hamster 65294. Am. J. Anat. 172, 317–330 [DOI] [PubMed] [Google Scholar]
- 15.Rejraji H., Sion B., Prensier G., Carreras M., Motta C., Frenoux J. M., Vericel E., Grizard G., Vernet P., Drevet J. R. (2006). Lipid remodeling of murine epididymosomes and spermatozoa during epididymal maturation. Biol. Reprod. 74, 1104–1113 [DOI] [PubMed] [Google Scholar]
- 16.Cooper T. G., Yeung C. H. (2010). Physiology of sperm maturation and fertilization. In Andrology: Male Reproductive Health and Dysfunction, 3rd ed.Nieschlag E., Behre H. M., Nieschlag S. (eds), Springer, Berlin [Google Scholar]
- 17.Saalmann A., Münz S., Ellerbrock K., Ivell R., Kirchhoff C. (2001) Novel sperm-binding proteins of epididymal origin contain four fibronectin type II-modules. Mol. Reprod. Dev. 58, 88–10 [DOI] [PubMed] [Google Scholar]
- 18.Kirchhoff C. (1999) Gene expression in the epididymis. Int. Rev. Cytol. 188, 133–202 [DOI] [PubMed] [Google Scholar]
- 19.Dacheux J. L., Belghazi M., Lanson Y., Dacheux F. (2006) Human epididymal secretome and proteome. Mol. Cell. Endocrinol. 250, 36–42 [DOI] [PubMed] [Google Scholar]
- 20.Sullivan R., Frenette G., Girouard J. (2007) Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J. Androl. 9, 483–491 [DOI] [PubMed] [Google Scholar]
- 21.Cooper T.G. (1998) Interactions between epididymal secretions and spermatozoa. J. Reprod. Fertil. Suppl. 53, 119–136 [PubMed] [Google Scholar]
- 22.Cooper T. G., Yeung C. H. (1999) Recent chemical approaches to post-testicular, epididymal contraception. Hum. Reprod. Update 5, 141–152 [DOI] [PubMed] [Google Scholar]
- 23.Li J. Y., Wang H. Y., Liu J., Liu Q., Zhang J. S., Wan F. C., Liu F. J., Jin S. H., Zhang Y. L. (2008) Transcriptome analysis of a cDNA library from adult human epididymis. DNA Res. 15, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thimon V., Koukoui O., Calvo E., Sullivan R. (2007) Region-specific gene expression profiling along the human epididymis. Mol. Hum. Reprod. 13, 691–704 [DOI] [PubMed] [Google Scholar]
- 25.Thimon V., Calvo E., Koukoui O., Légaré C., Sullivan R. (2008) Effects of vasectomy on gene expression profiling along the human epididymis. Biol. Reprod. 79, 262–273 [DOI] [PubMed] [Google Scholar]
- 26.Dacheux J. L., Gatti J. L., Dacheux F. (2003) Contribution of epididymal secretory proteins for spermatozoa maturation. Microsc. Res. Tech. 61, 7–17 [DOI] [PubMed] [Google Scholar]
- 27.Dacheux J. L., Belleannée C., Jones R., Labas V., Belghazi M., Guyonnet B., Druart X., Gatti J. L., Dacheux F. (2009) Mammalian epididymal proteome. Mol. Cell. Endocrinol. 306, 45–50 [DOI] [PubMed] [Google Scholar]
- 28.WHO (1999) World Health Organization Laboratory Handbook for the examination of human semen and sperm-cervical interaction. CUP Press, Cambridge, UK [Google Scholar]
- 29.Cooper T. G., Keck C., Oberdieck U., Nieschlag E. (1993) Effects of multiple ejaculations after extended periods of sexual abstinence on total, motile and normal sperm numbers, as well as accessory gland secretions from healthy normal and oligozoospermic men. Hum. Reprod. 8, 1251–1258 [DOI] [PubMed] [Google Scholar]
- 30.Jennings M. G., Temple-Smith P. D., Southwick G. J,, Nauadu P. L., Baker H. W. (1987) Patterns of human epididymal fluid proteins on polyacrylamide gel electrophoresis. Int. J. Androl. 10, 441–446 [DOI] [PubMed] [Google Scholar]
- 31.Yeung C. H., Cooper T. G., Bergmann M., Schulze H. (1991) Organization of tubules in the human caput epididymidis and the ultrastructure of their epithelia. Am. J. Anat. 191, 261–279 [DOI] [PubMed] [Google Scholar]
- 32.Gotham S. M., Fryer P. J., Paterson W. R. (1988) The measurement of insoluble proteins using a modified Bradford assay. Anal. Biochem. 173, 353–358 [DOI] [PubMed] [Google Scholar]
- 33.Görg A., Weiss W., Dunn M. J. (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 34.Kumarathasan P., Mohottalage S., Goegan P., Vincent R. (2005). An optimized protein in-gel digest method for reliable proteome characterization by MALDI-TOF-MS analysis. Anal. Biochem. 346, 85–89 [DOI] [PubMed] [Google Scholar]
- 35.Rajcevic U., Petersen K., Knol J. C., Loos M., Bougnaud S., Klychnikov O., Li K. W., Pham T. V., Wang J., Miletic H., Peng Z., Bjerkvig R., Jimenez C. R., Niclou S. P. (2009) iTRAQ based proteomic profiling reveals increased metabolic activity and cellular crosstalk in angiogenic compared to invasive Glioblastoma phenotype. Mol. Cell. Proteomics. 8, 2595–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommerer N., Centeno D., Rossignol M. (2007) Peptide mass fingerprinting: identification of proteins by MALDI-TOF. Methods Mol. Biol. 355, 219–234 [DOI] [PubMed] [Google Scholar]
- 37.Lin Y. Q., Li J. Y., Wang H. Y., Liu J., Zhang C. L., Wang W. T., Liu J., Li N., Jin S. H. (2008) Cloning and identification of a novel sperm binding protein, HEL-75, with antibacterial activity and expressed in the human epididymis. Hum. Reprod. 23, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 38.Cheng G. Z., Li J. Y., Li F., Wang H. Y., Shi G. X. (2009). Human ribonuclease 9, a member of ribonuclease A superfamily, specifically expressed in epididymis, is a novel sperm-binding protein. Asian J. Androl. 11, 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cyranoski D. (2003) China takes centre stage for liver proteome. Nature 425, 441. [DOI] [PubMed] [Google Scholar]
- 40.Service R. F. (2003) Proteomics. Public projects gear up to chart the protein landscape. Science 302, 1316–1318 [DOI] [PubMed] [Google Scholar]
- 41.Chinese Human Liver Proteome Profiling Consortium (2010) First Insight into the Human Liver Proteome from PROTEOME(SKY)-LIVER(Hu) 1.0, a Publicly Available Database. J. Proteome Res. 9, 79–94 [DOI] [PubMed] [Google Scholar]
- 42.Baker M. A., Hetherington L., Reeves G. M., Aitken R. J. (2008) The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 8, 1720–1730 [DOI] [PubMed] [Google Scholar]
- 43.Baker M. A., Hetherington L., Reeves G. M., Müller J., Aitken R. J. (2008). The rat sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 8, 2312–2321 [DOI] [PubMed] [Google Scholar]
- 44.Johnston D. S., Wooters J., Kopf G. S., Qiu Y., Roberts K. P. (2005) Analysis of the human sperm proteome. Ann. N.Y. Acad. Sci. 1061, 190–202 [DOI] [PubMed] [Google Scholar]
- 45.Martínez-Heredia J., Estanyol J. M., Ballescà J. L., Oliva R. (2006) Proteomic identification of human sperm proteins. Proteomics 6, 4356–4369 [DOI] [PubMed] [Google Scholar]
- 46.Oliva R., Martínez-Heredia J., Estanol J. M. (2008) Proteomics in the study of the sperm cell composition, differentiation and function. Syst. Biol. Reprod. Med. 5, 23–36 [DOI] [PubMed] [Google Scholar]
- 47.Oliva R., de Mateo S., Estanyol J. M. (2009) Sperm cell proteomics. Proteomics 9, 1004–1017 [DOI] [PubMed] [Google Scholar]
- 48.Baker M. A., Aitken R. J. (2009) Proteomic insights into spermatozoa: critiques, comments and concerns. Expert Rev. Proteomics 6, 691–705 [DOI] [PubMed] [Google Scholar]
- 49.Sullivan R., Frenette G., Girouard J. (2007) Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J. Androl. 9, 483–491 [DOI] [PubMed] [Google Scholar]
- 50.Thimon V., Frenette G., Saez F., Thabet M., Sullivan R. (2008) Protein composition of human epididymosomes collected during surgical vasectomy reversal: a proteomic and genomic approach. Hum. Reprod. 23, 1698–1707 [DOI] [PubMed] [Google Scholar]
- 51.Baker M. A., Witherdin R., Hetherington L., Cunningham-Smith L., Aitken R. J. (2005) Identification and post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics 5, 1003–1012 [DOI] [PubMed] [Google Scholar]
- 52.Jelinsky S. A., Turner T. T., Bang H. J., Finger J. N., Solarz M. K., Wilson E., Brown E. L., Kopf G. S., Johnston D. S. (2007) The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol. Reprod. 76, 561–570 [DOI] [PubMed] [Google Scholar]
- 53.Lin Y. Q., Li J. Y., Wang H. Y., Liu J., Zhang C. L., Wang W. T., Liu J., Li N., Jin S. H. (2008) Cloning and identification of a novel sperm binding protein, HEL-75, with antibacterial activity and expressed in the human epididymis. Hum. Reprod. 23, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 54.Cornwall G. A. (2009) New insights into epididymal biology and function. Hum. Reprod. Update 15, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooper T. G. (1998). Epididymis. In Encyclopedia of Reproduction. Neill J. D., Knobil E., (eds.), Academic, San Diego, CA, Vol. 2, pp. 1–17 [Google Scholar]
- 56.Baldi E., Luconi M., Bonaccorsi L., Forti G. (2002) Signal transduction pathways in human spermatozoa. J. Reprod. Immunol. 53, 121–131 [DOI] [PubMed] [Google Scholar]
- 57.Zhang J., Wu J., Huo R., Mao Y., Lu Y., Guo X., Liu J., Zhou Z., Huang X., Sha J. (2007) ERp57 is a potential biomarker for human fertilization capability. Mol. Hum. Reprod. 13, 633–639 [DOI] [PubMed] [Google Scholar]
- 58.Lefèvre A., Martin Ruiz C., Chokomian S., Duquenne C., Finaz C. (1997) Characterization and isolation of SOB2, a human sperm protein with a potential role in oocyte membrane binding. Mol. Hum. Reprod. 3, 507–516 [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Zhang N., Zhang X., Miao S., Zong S., Koide S. S., Wang L. (2009) Experimental immunological infertility effect of anti-GAPDH-2 antibodies on the fertility of female mice. Fertil. Steril. 92, 2020–2027 [DOI] [PubMed] [Google Scholar]
- 60.Wolkowicz M. J., Digilio L., Klotz K., Shetty J., Flickinger C. J., Herr J. C. (2008) Equatorial segment protein (ESP) is a human Alloantigen involved in sperm-egg binding and fusion. J. Androl. 29, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.