Abstract
Microarray-based sandwich immunoassays can simultaneously detect dozens of proteins. However, their use in quantifying large numbers of proteins is hampered by cross-reactivity and incompatibilities caused by the immunoassays themselves. Sequential multiplex analyte capturing addresses these problems by repeatedly probing the same sample with different sets of antibody-coated, magnetic suspension bead arrays. As a miniaturized immunoassay format, suspension bead array-based assays fulfill the criteria of the ambient analyte theory, and our experiments reveal that the analyte concentrations are not significantly changed. The value of sequential multiplex analyte capturing was demonstrated by probing tumor cell line lysates for the abundance of seven different receptor tyrosine kinases and their degree of phosphorylation and by measuring the complex phosphorylation pattern of the epidermal growth factor receptor in the same sample from the same cavity.
Phosphorylation of proteins is an integral part of the signal transduction of eukaryotic cells as it modulates the activity of complex protein networks. Although Western blot- and immunoprecipitation-based MS approaches (1, 2) can lead to detailed insights into these processes, most of the integrated approaches only allow a static view of protein phosphorylation because they are not suitable for the screening of hundreds of samples. Either planar or bead array-based sandwich immunoassays can be used to analyze the quantity and activation state of signaling molecules in multiplex, enabling the systematic profiling of protein abundance and post-translational modifications (3–6) in hundreds of samples. However, multiplex immunoassays are only suitable for the simultaneous analysis of a limited number of proteins. The detection of comprehensive phosphorylation patterns is difficult as this involves assay systems that are incompatible with multiplexing.
In principle, two sandwich immunoassay setups are possible for probing the phosphorylation state of a protein. The first setup applies a capture antibody specific for a non-modified part of the protein and uses a phosphorylation state-specific detection antibody. When applied to an array-based format, however, this setup does not allow for the simultaneous measurement of the abundance and the degree of phosphorylation (3, 4). A mixture of detection antibodies, one specific for the phosphorylation site and one specific for the non-modified site of the protein, would bind simultaneously to the two different epitopes, and assay signals could not be further deconvoluted by the spatial or color code of the array. The second sandwich immunoassay setup for the analysis of protein phosphorylation applies a phosphorylation state-specific capture antibody and a protein-specific detection antibody. In such a setup, an anti-phosphotyrosine antibody (e.g. mAb 4G10) cannot be applied as a capture antibody because a huge variety of tyrosine phosphorylated proteins would be captured, and specific signals could rarely be deconvoluted. Using capture antibodies that bind to phosphorylated epitopes in the context of their flanking amino acids is not a problem until a multiplex readout is desired. If one antibody specific for the phosphosite and one antibody specific for the abundance of a protein are used together in a multiplex assay panel they might compete for their analyte. The situation becomes even more complex if the protein of interest contains various phosphorylation sites such as e.g. the epidermal growth factor receptor. Several capture antibodies target different epitopes of the same protein and therefore compete for the overall amount of targeted protein in the sample, thus making a valid simultaneous measurement problematic.
Although different ways of tackling the problem of assay multiplexing are in use, we demonstrated the feasibility to sequentially perform such incompatible assays from the same sample using a magnetic particle handler that moves particles through the samples and reagents (Fig. 1). Using a model assay, we confirmed that suspension bead array-based immunoassays work under ambient analyte conditions. As described by Roger Ekins (7), decreasing of the amount of capture antibody in a sandwich immunoassay setup from a macrospot (e.g. a microtiter plate assay) to a microspot generates a scenario where only a tiny fraction of the present target analytes is captured on the microspot. Therefore, the overall concentration of the analyte molecules in the sample does not change significantly even in the case of low target concentrations and high affinity binding reactions. Furthermore, as the initial concentration of the analyte is not significantly changed when performing a miniaturized sandwich immunoassay, multiple post-translational modifications within the same protein can be measured either in sequence or in parallel in the same multiplex panel.
Fig. 1.
Sequential multiplex analyte capturing. Magnetic suspension bead array assays can be performed sequentially, reusing the same sample material (indicated by the blue arrow). The use of a magnetic particle handler enables the quantitative transfer (black arrow) of the magnetic beads from the sample well into the wells containing washing solutions or other assay reagents. Magnetic beads from the first bead array panel are incubated with the samples to capture their respective analyte. Then the magnetic beads are subjected to washing and detection steps and are finally transferred into the readout plate (first row). After retracting the magnetic suspension bead array of the first assay panel from the sample, a bead array from the second assay panel is added and processed as described above but using different detection antibodies (second row). A third bead array assay panel can be applied after removing the second panel (third row) and so on.
By probing tumor cell lines for the abundance of seven different receptor tyrosine kinases and their generic tyrosine phosphorylation, we generated complex phosphorylation patterns and thereby demonstrated the potential of this approach. More importantly, demonstrating ambient analyte conditions allowed the parallel detection of phosphorylation at different sites of the EGFR1 using phosphorylation site-specific antibodies as capture molecules with one assay panel. Phosphorylation of eight different sites and the abundance of the EGFR could be quantified relative to one another without any interference of the different immunoassays during multiplexing because competition for the analyte can be prevented by running the assays under ambient analyte conditions.
EXPERIMENTAL PROCEDURES
Cell Culture Treatment
106 A431 cells were seeded in T75 flasks and cultured in DMEM (Invitrogen) supplemented with 2 mm glutamine and 10% fetal calf serum (Invitrogen) until 80% confluence. Cells were cultured in 5% CO2 atmosphere and at 37 °C. 16 h before treatment cultures were starved in culture medium lacking fetal calf serum. Epidermal growth factor (EMD Chemicals) was dissolved 10 mm acetic acid (EMD Chemicals) and 0.1% bovine serum albumin (EMD Chemicals) to give a concentration of 100 μg/ml. For stimulation experiments, the growth factor was diluted 1:1000 in starving medium and applied for 10 min. The phorbol ester phorbol 12-myristate 13-acetate (PMA; EMD Chemicals) was dissolved at a concentration of 10 mg/ml in DMSO (EMD Chemicals). Cells were treated for 60 min with a concentration of 100 ng/ml in culture medium lacking fetal calf serum. Pervanadate (Merck KGaA) and H2O2 (Merck KGaA) were applied to the cells in a concentration of 10 mm for 10 min.
Preparation of Cell Lysates
After treatments, cells were rinsed two times with ice-cold Tris-buffered saline (Sigma-Aldrich). Proteins were extracted using a cell extraction kit (EMD Chemicals). Extraction buffer (2 ml) freshly supplemented with phosphatase inhibitor mixture set V (EMD Chemicals), protease inhibitor mixture set III (EMD Chemicals), and Benzonase (EMD Chemicals) was applied per cell culture dish. Sample preparation was completed after 20 min of gentle rocking at 4 °C. Cell debris were removed using a centrifuge (Eppendorf) at a speed of 10,000 relative centrifugal force at 4 °C. Protein concentration was determined using a BCA protein assay (Thermo Scientific).
Antibody Coupling
Capture antibody goat anti-biotin (Sigma-Aldrich) was covalently coupled to magnetic carboxylated fluorescent beads (Luminex Corp.) according to the manufacturer's protocol. In brief, 200 μl of each bead set (2.5 × 106 magnetic beads) were captured for 2 min using a magnetic bead separator (Invitrogen), and the supernatant was removed. The beads were washed twice with 100 μl of activation buffer (0.1 m Na2HPO4, pH 6.2; EMD Chemicals) and resuspended in 80 μl of the same buffer. Beads were activated by adding freshly prepared solutions of N-hydroxysulfosuccinimide sodium salt (50 mg/ml; Thermo Scientific) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (50 mg/ml; Thermo Scientific) in activation buffer. 10 μl of each solution were added to each reaction and kept for 20 min at room temperature in the dark while rotating. Subsequently, the microspheres were washed twice with 500 μl of coupling buffer (50 mm 2-morpholinoethanesulfonic acid, pH 5.0; Sigma-Aldrich) followed by adding 12.5 μg of antibody (50 μg/ml) in 250 μl of the same buffer. The suspension was incubated on a shaker for 2 h at room temperature in the dark. Excess protein was removed by two washing steps using 1 ml of washing buffer (0.1% Tween (v/v) in PBS, pH 7.4). Finally, microspheres were resuspended in 100 μl of PBS containing 1% BSA (w/v) (Carl Roth), 0.05% NaN3 (w/v) (Sigma-Aldrich) and stored in the dark at 2–8 °C.
Biotinylated R-phycoerythrin (Biotin-PE) Immunoassay
Biotin-PE (Invitrogen) was diluted to concentrations of 300, 80, 20, 5, and 1 pm in Blocking reagent for ELISA (Roche Applied Science). 100 μl of each solution were pipetted into a PCR microtiter plate (Abgene). Magnetic beads coated with a goat anti-biotin IgG were added to these samples using a magnetic particle handler (KingFisher 96, Thermo Scientific). Beads were incubated for a 30-min period at RT followed by a collection and transfer step to a second microtiter plate (Abgene) for the readout in a Luminex instrument. The assay was performed five times from the same sample. Parameters of the magnetic bead handler protocol for the biotin-PE immunoassay can be found in Table I. Samples were analyzed in triplicates. The data were acquired with a Luminex 100 instrument (Luminex Corp.) according to the manufacturer's instructions.
Table I. Parameters of magnetic bead handler protocol for biotin-PE immunoassay.
| Step | Plate content | Volume | Mixing parameters |
||
|---|---|---|---|---|---|
| Time | Temperature | Speed | |||
| μl | |||||
| 1) Transfer | Magnetic bead array | 50 | |||
| 2) Analyte capture | Sample | 100 | 30 min | RT | 750 rpm |
| 3) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 4) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 5) Bead release to assay plate | Assay buffer | 100 | 20 s | RT | Slow |
Sequential Multiplex Analyte Capturing: Receptor Tyrosine Kinase Profiling
Three different bead-based assay kits for receptor tyrosine kinase profiling were used for the sequential multiplex analyte capturing experiment: (i) WidescreenTM site-specific phospho-EGFR panel, (ii) Widescreen receptor tyrosine kinase panel for assaying generic tyrosine phosphorylation, and (iii) Widescreen receptor tyrosine kinase panel for the quantification of the seven different receptor tyrosine kinases (EMD Chemicals). Assays were performed sequentially as follows. All assay steps besides the first incubation step were done using a magnetic particle handler (KingFisher 96, Thermo Scientific). All reagents including wash buffer, detection antibody mixture, and streptavidin-phycoerythrin were prediluted as recommended by the manufacturer and placed in the magnetic particle handler. Protein concentration of the extract was adjusted to 1 mg/ml in lysis buffer (EMD Chemicals). Subsequently, samples were diluted to a concentration of 0.1 μg/ml using assay diluent (EMD Chemicals) and pipetted into a 96-well PCR plate (Abgene). Magnetic beads of the first assay panel were transferred into the samples using the magnetic particle handler. Protein extracts were incubated with the suspension bead arrays overnight at 8 °C in a 96-well PCR plate while shaking (750 rpm; Eppendorf) outside the magnetic particle robot. After the first incubation step, magnetic beads were transferred from the assay plate to wash plates, and the bead array of the following assay panel was added by the magnetic particle handler. Incubation of the first array panel with the detection antibody mixture was carried out for 60 min and with streptavidin-phycoerythrin conjugate as described in Table II. The second and third assay panels were processed as described above 24 and 48 h later. The data were acquired with a Luminex 100 instrument (Luminex Corp.) according to the manufacturer's instructions. Samples were analyzed in triplicates. The background readings for each capture antibody were obtained by incubating the bead panels with 10% cell extraction kit (EMD Chemicals). Parameters of the magnetic bead handler protocol for the sandwich immunoassay can be found in Table II.
Table II. Parameters of magnetic bead handler protocol for sandwich immunoassay.
| Step | Plate content | Volume | Mixing parameters |
||
|---|---|---|---|---|---|
| Time | Temperature | Speed | |||
| μl | |||||
| 1) Transfer | Magnetic bead array | 50 | |||
| 2) Analyte capture | Crude cell culture lysate | 100 | Overnight | 8 °C | 750 rpm |
| 3) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 4) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 5) Detection antibody (5 cycles) | Detection antibody mixture | 100 | 10 min, pause | ||
| 2 min | RT | Very slow | |||
| 6) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 7) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 8) Detection reagent (4 cycles) | SAPE solution | 100 | 10 min, pause | ||
| 2 min | RT | Very slow | |||
| 9) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 10) Wash + transfer | Wash buffer | 100 | 20 s | RT | Slow |
| 11) Bead release to assay plate | Assay buffer | 100 | 20 s | RT | Slow |
RESULTS
Measurements under Ambient Analyte Conditions
The “ambient analyte theory,” as coined by Roger Ekins (7), states that only a small fraction of analyte is captured from the sample when using a microarray-based assay system. As a result, the concentration of analyte is not significantly changed by the assay itself even at low analyte concentrations and for high affinity binding reactions. This implies that the same sample can be analyzed more than once for the same analyte as no depletion should occur when a miniaturized assay system is applied. This should therefore enable the simultaneous detection of different phosphorylations in the same assay panel or allow the multiplex detection of the different modification states of a protein in a sequential mode without any interference that might arise from different analyte variants competing for capture proteins.
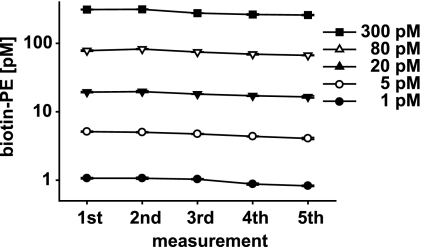
We proved ambient analyte conditions for suspension bead arrays by repeatedly probing the same sample for the same analyte. A magnetic particle handler involving 96 magnetic rods was used to move the antibody-coated particles through the samples and reagents. A model assay involving an anti-biotin antibody as capture molecule and a biotin-PE analyte was used to assess the validity of ambient analyte conditions in suspension bead arrays. Magnetic beads coated with goat anti-biotin IgG were used to capture the biotin-PE in five sequential assays (Fig. 2). The beads were sequentially added to the sample containing different analyte concentrations (300, 80, 20, 5, and 1 pm), incubated for 30 min, and transferred to a second microtiter plate for subsequent analysis with a flow cytometer-like system (Luminex 100). The signal intensity of the biotin-PE analyte remained virtually unchanged in all five assays (Fig. 2). The overall signal intensity decrease after each assay was less than 5% (Table III), which is in line with the ambient analyte theory of Ekins (7).
Fig. 2.
Ambient analyte theory: repetitive measurement of biotin-PE from same sample. The diagram shows the results for five sequentially performed analyses of the same protein from the same sample from the same well. A model assay using biotin-PE and a biotin-specific capture antibody was used. Anti-biotin antibody-coated magnetic beads were incubated with biotin-PE at five different concentrations (300, 80, 20, 5, and 1 pm) in triplicates. After a 30-min incubation period, the beads were removed from the sample, and new antibody-coupled magnetic beads were added. This procedure was repeated four times, and the assay was read out on a Luminex 100 reader. No analyte limitations were detectable over the five measurements. The data from the first measurement were used for a five-parametric logistic fit. Using the parameters of this fit, the raw assay data were transformed to analyte concentrations (see Table III). After five assays, an analyte decrease of 20% was observed in total, suggesting a captured amount of less than 5% per assay.
Table III. Ambient analyte theory: repetitive measurement of biotin-PE from same sample.
Shown are the results for five sequentially performed analyses of biotin-PE from the same sample from the same well. The data from the first measurement were used for a five-parametric logistic fit. Using the parameters of this fit, the raw assay data were transformed to analyte concentrations. After five assays, an analyte decrease of 20% was observed in total, suggesting a captured amount of less than 5% per assay. AU, arbitrary units.
| Biotin-PE | First measurement |
Second measurement |
Third measurement |
Fourth measurement |
Fifth measurement |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | Mean | %CV | |
| pm | AU | AU | AU | AU | AU | |||||
| Blank | 8 | 8 | 8 | 16 | 8 | 8 | 8 | 13 | 8 | 13 |
| 1 | 82 | 2 | 82 | 3 | 80 | 1 | 72 | 3 | 70 | 2 |
| 5 | 315 | 2 | 309 | 1 | 293 | 1 | 271 | 1 | 253 | 3 |
| 20 | 1139 | 2 | 1158 | 3 | 1071 | 0 | 1015 | 1 | 981 | 3 |
| 80 | 3466 | 2 | 3580 | 1 | 3366 | 1 | 3211 | 2 | 3131 | 1 |
| 300 | 6181 | 1 | 6191 | 1 | 5996 | 0 | 5926 | 2 | 5895 | 1 |
| pm | %CV | pm | %CV | pm | %CV | pm | %CV | pm | %CV | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 2 | 1 | 3 | 1 | 1 | 1 | 3 | 1 | 2 |
| 5 | 5 | 2 | 5 | 1 | 5 | 1 | 4 | 1 | 4 | 3 |
| 20 | 19 | 2 | 20 | 3 | 18 | 0 | 17 | 1 | 17 | 3 |
| 80 | 78 | 2 | 83 | 1 | 75 | 1 | 69 | 2 | 67 | 1 |
| 300 | 312 | 1 | 314 | 1 | 276 | 0 | 265 | 2 | 260 | 1 |
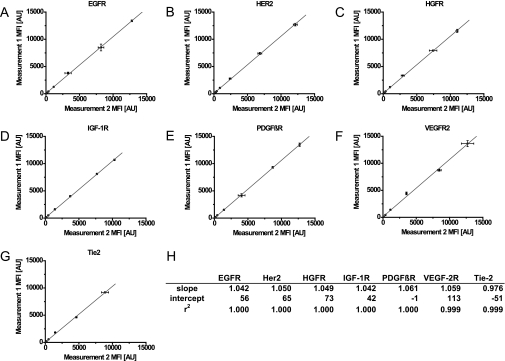
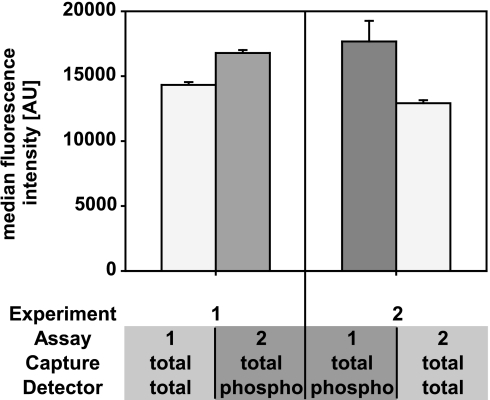
Based on these results, the quantities of seven recombinant receptor tyrosine kinase standards were measured. Two measurements were carried out from the same standard dilution curve on 2 separate days. Color-coded magnetic beads coated with capture antibodies that are specific for different RTKs (EGFR, HER2, HGFR, IGF-1R, PDGFRβ, VEGFR2, and Tie-2) were incubated overnight with different concentrations of standard recombinant receptor proteins. The beads were subsequently removed with the magnetic particle handler, and a new batch of color-coded magnetic beads coated with the same capture antibodies was added to the same RTK solutions and incubated overnight. The beads were washed and incubated with the respective biotinylated detection antibodies. Streptavidin-conjugated R-phycoerythrin (SAPE) was used as a reporter molecule. No significant differences were observed between the first and second measurements (Fig. 3, A–G). For EGFR, a correlation of r2 = 1.000 and a deviation of 0.042 from the optimal slope 1 was calculated (Fig. 3, A and H). Experimental results for the other RTKs gave similar correlations and values (Fig. 3H). It can therefore be assumed that sequential measurements of the same analyte from the same sample do not lead to signal intensity losses resulting from sample depletion or protein denaturation. A sequential assessment was subsequently made of the presence of EGFR and the degree of tyrosine phosphorylation in a protein extract from an epidermal growth factor (EGF)-stimulated A431 cell culture by comparing the order in which the two assays were carried out. In one assay, the quantification of EGFR was followed by the quantification of phosphorylated EGFR, and in the other, the quantification of phosphorylated EGFR was followed by the quantification of EGFR. Only slight differences were found regardless of whether the phosphorylation or the total amount of protein was measured first or second (Fig. 4). These experiments established the basis for performing (a) a sequential assessment of the modification of a protein and its absolute amount and (b) a parallel analysis of the modification of a protein and its concentration in the same sample.
Fig. 3.
Repetitive measurements of seven receptor tyrosine kinases from same sample. The diagrams show the results of two sequentially conducted multiplex sandwich immunoassays for the EGFR (A), HER2 (B), HGFR (C), IGF-1R (D), PDGFRβ (E), VEGFR2 (F), and Tie-2 (G) using the same dilution series. Antibody-coupled beads were incubated with recombinant standards at seven different 4-fold dilutions starting for EGFR, HER2, and PDGFβR at 20,000 pg/ml; for IGF-1R and VEGFR at 100,000 pg/ml; for HGFR at 50,000 pg/ml; and for Tie-2 at 10,000 pg/ml. A mixture of biotinylated antibodies specific for the respective RTK, together with SAPE, was used to detect the analytes in the readout system (Luminex 100). The median fluorescence intensities (MFI) of the two sequential assays were plotted against each other and fitted linearly. Standard deviations were calculated from three technical replicates. The table (H) contains slopes, intercepts, and correlation coefficients of the fits for the seven assays. The calculated correlations and slopes close to 1.000 confirm the hypothesis that the analyte can be analyzed twice without changing the concentration of the analyte itself. AU, arbitrary units.
Fig. 4.
Sequential measurement of EGFR and tyrosine phosphorylation of EGFR from same sample. An EGF-stimulated A431 cell line was used for the sequential measurement of EGFR and the generic tyrosine phosphorylation of EGFR. The median fluorescence intensities (n = 3) obtained in two experiments are plotted in the diagram. The left panel shows the results for the experimental order EGFR followed by tyrosine phosphorylated EGFR measurement, and the right panel shows the inverted order. The results do not differ significantly. AU, arbitrary units.
Sequential Multiplex Analyte Capturing: Receptor Tyrosine Kinase Profiling
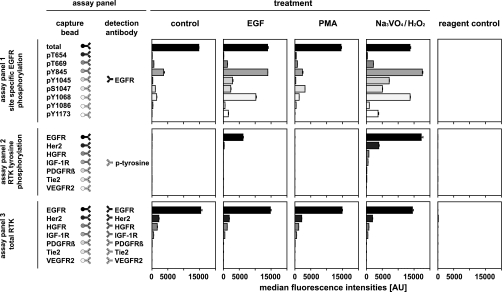
Consequently, different multiplex sandwich immunoassays were performed using the same sample three times sequentially. Lysates from A431 cells treated with either EGF, PMA, or a combination of sodium orthovanadate and hydrogen peroxide were probed with three different bead array sandwich immunoassay panels. Fig. 5 shows the results and a schematic of (i) a 9-plex sandwich immunoassay used for the analysis of eight EGFR phosphorylation sites, (ii) a 7-plex sandwich immunoassay used to analyze the generic tyrosine phosphorylation of seven different RTKs, and (iii) a 7-plex sandwich immunoassay used to measure the relative quantities of the seven RTKs. As expected, the amount of the four RTKs EGFR, HER2, HGFR, and IGF-1R did not change in the differentially treated cells. The receptors PDGFRβ, Tie-2, and VEGFR2 could not be detected. EGF stimulation led to a generic increase in tyrosine phosphorylation of EGFR and a slight increase in tyrosine phosphorylation of HER2 (8). The EGFR phosphorylation status examined with a 9-plex assay confirmed previously published results: phosphorylation at tyrosine 1045 (9); tyrosines 1068, 1086, and 1173 (9–11); threonine 669 (12, 13); serine 1047 (14); and tyrosine 845 (13, 15). PMA stimulation showed a higher degree of phosphorylation of threonine 654 (16) and serine 1047 (14). The treatment of the A431 cells with the phosphatase inhibitor sodium orthovanadate and reactive oxygen species resulted in an elevated phosphorylation of all phosphosites investigated.
Fig. 5.
Phosphorylation analysis of treated A431 cell line using sequential multiplex analyte capturing. A431 cell cultures were treated with EGF, PMA, and a combination of sodium vanadate and hydrogen peroxide to give different responses in receptor signaling. First, nine different color-coded beads (assay panel 1) conjugated with antibodies specific for total EGFR and EGFR phosphorylation at threonines 654 and 669; serine 1047; and tyrosines 845, 1045, 1068, 1086, and 1173 were incubated with the protein extract sample overnight. To carry out the detection, the beads were then removed from the sample using a magnetic bead handling robot and added to a solution of biotinylated EGFR-specific antibody followed by a third incubation step with SAPE. After removing the beads of panel 1 from the sample, antibody-coupled beads from assay panel 2 (seven RTKs: EGFR, HER2, HGFR, IGF-1R, PDGFRβ, VEGFR2, and Tie-2) were added and incubated with the sample overnight. Generic phosphorylation was detected by retracting the beads from the sample followed by incubation with a biotinylated tyrosine phosphorylation-specific antibody and SAPE. In the final analysis, the antibody-coupled beads from assay panel 3 were added to the same samples and incubated overnight. The relative abundance of the RTKs was detected by incubation with an antibody mixture of seven different biotinylated antibodies specific for the respective RTK. The figure shows the median fluorescence intensities (n = 3) for the sequentially performed assay panels.
DISCUSSION
A key driver for the development of multiplexed immunoassays is the demand for detailed information on the amount and on the modification/activation state of a multitude of proteins. With the growing knowledge on cellular signaling processes, a more detailed understanding of their complexity has been obtained (17–19). Signaling cascades have been elucidated, and the cross-talk between different pathways has been described, and it is now clear that the description of signaling processes requires information on a large number of proteins and on their activation state (20, 21). Here classical approaches like the Western blot, microtiter plate-based immunoassays, and mass spectrometry-based methods show their limitations because the workload and the required amount of material exceed the possibilities of a single laboratory (18, 22). Miniaturization and parallelization of immunoassays address these problems, and protein microarrays using beads and planar surfaces that allow the sensitive detection of proteins from a low amount of sample material are available (2–4, 23). Nevertheless, limitations caused by the cross-reactivity of antibodies and the availability of matched antibody pairs often make multiplexing of sandwich immunoassays difficult (24, 25). Here we show that the use of paramagnetic beads for suspension bead array-based immunoassays opens the possibility to sequentially run multiplexed assays from the very same sample, and this approach allows circumventing incompatibility or cross-reactivity problems of antibodies. For the strategy used, smaller sets of compatible assay panels are defined and performed in sequence from the same initial sample. The use of magnetic bead handlers makes this approach feasible, and although the degree of multiplexing within one measurement is lower, the repeated measurements keep the advantages of assay multiplexing. To test the value of this strategy, we first decided to look at the ambient analyte theory as described by Ekins (7). Ekins showed that the maximum sensitivity in a miniaturized assay system is obtained when the capturing process does not change the analyte concentration in the sample. This also means that repeated measurements under these conditions can be performed without resulting in significant changes in analyte concentration in the sample. Using a simple model assay, it was shown that the measured concentration of analyte was not changed in five repetitive measurements (Fig. 2). This shows that the ambient analyte theory that rules miniaturized assay systems is applicable and that the small amount of capture antibody used in a suspension bead array causes a negligible depletion of analyte in the sample.
As mentioned above, profiling of signaling pathways requires the determination of protein phosphorylation as a surrogate for protein activity or inactivity (e.g. kinase). The determination of the absolute amount of a protein and its phosphorylation state is often not possible within one multiplex immunoassay. The determination of multiple phosphorylation sites therefore requires independent measurements because antibodies used for one sandwich assay will interfere with antibodies from another sandwich immunoassay.
Here the sequential multiple analyte capture concept shows its value. The system allows the analysis of multiple phosphorylations of receptor tyrosine kinases from the very same sample by sequential measurements, but no significant changes in analyte concentration after each of the sequential measurements are observed. This was tested in a sequential analysis of a protein extract prepared from an EGF-stimulated cell line as shown in Fig. 4. The analysis of differentially treated cell culture samples was performed, and the degree of phosphorylation of seven receptor tyrosine kinases and eight known phosphorylation sites of the epidermal growth factor receptor were measured. Treatment of the cell line A431 with EGF, PMA, and oxidative stress led to the detection of the changes in EGFR and RTK phosphorylation that have been described in the literature (8–16). However, our work flow allowed the generation of a picture of the changes in the phosphorylation patterns using 20 times less sample material and a fraction of hands-on time. This concept is therefore of special interest if limited material like biopsies or laser capture-microdissected sample material has to be analyzed. The sequential use of different array-based assays should allow gathering information of up to 50 different proteins or protein modifications requiring only 10 μg of protein extracts. In addition, the approach provides a practical workaround of some of the practical limitations of multiplexing namely assay incompatibilities and the cross-reactivity of the antibodies.
Acknowledgments
We kindly thank Jutta Bachmann for proofreading the manuscript. Thomas Joos is a member of the scientific advisory board of Luminex Corp., Austin, TX. Tanja Henzler and Thomas Herget are employees of Merck KGaA, Darmstadt, Germany. They develop and commercialize bead-based multiplexed assays.
Footnotes
* This work was supported by German Federal Ministry of Education (Bundesministerium für Bildung und Forschung) Grant FKZ 313081E (Systems Biology).
1 The abbreviations used are:
- EGFR
- epidermal growth factor receptor
- HER2
- human epidermal growth factor receptor 2
- HGFR
- hepatocyte growth factor receptor
- IGF-1R
- insulin-like growth factor 1 receptor
- PDGFRβ
- platelet-derived growth factor receptor β
- PMA
- phorbol 12-myristate 13-acetate
- RTK
- receptor tyrosine kinase
- SAPE
- streptavidin-conjugated R-phycoerythrin
- Tie-2
- tyrosine kinase with immunoglobulin and EGF repeats 2
- VEGFR2
- vascular endothelial growth factor receptor 2
- biotin-PE
- biotinylated R-phycoerythrin.
REFERENCES
- 1.Schmelzle K., White F. M. (2006) Phosphoproteomic approaches to elucidate cellular signaling networks. Curr. Opin. Biotechnol. 17, 406–414 [DOI] [PubMed] [Google Scholar]
- 2.Gaudet S., Janes K. A., Albeck J. G., Pace E. A., Lauffenburger D. A., Sorger P. K. (2005) A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol. Cell. Proteomics 4, 1569–1590 [DOI] [PubMed] [Google Scholar]
- 3.Khan I. H., Mendoza S., Rhyne P., Ziman M., Tuscano J., Eisinger D., Kung H. J., Luciw P. A. (2006) Multiplex analysis of intracellular signaling pathways in lymphoid cells by microbead suspension arrays. Mol. Cell. Proteomics 5, 758–768 [DOI] [PubMed] [Google Scholar]
- 4.Du J., Bernasconi P., Clauser K. R., Mani D. R., Finn S. P., Beroukhim R., Burns M., Julian B., Peng X. P., Hieronymus H., Maglathlin R. L., Lewis T. A., Liau L. M., Nghiemphu P., Mellinghoff I. K., Louis D. N., Loda M., Carr S. A., Kung A. L., Golub T. R. (2009) Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat. Biotechnol. 27, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paweletz C. P., Charboneau L., Bichsel V. E., Simone N. L., Chen T., Gillespie J. W., Emmert-Buck M. R., Roth M. J., Petricoin E. F., 3rd, Liotta L. A. (2001) Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 6.Nielsen U. B., Cardone M. H., Sinskey A. J., MacBeath G., Sorger P. K. (2003) Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc. Natl. Acad. Sci. U.S.A. 100, 9330–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekins R. P. (1989) Multi-analyte immunoassay. J. Pharm. Biomed. Anal. 7, 155–168 [DOI] [PubMed] [Google Scholar]
- 8.Wolf-Yadlin A., Kumar N., Zhang Y., Hautaniemi S., Zaman M., Kim H. D., Grantcharova V., Lauffenburger D. A., White F. M. (2006) Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol. 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 10.Downward J., Parker P., Waterfield M. D. (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485 [DOI] [PubMed] [Google Scholar]
- 11.Hsuan J. J., Totty N., Waterfield M. D. (1989) Identification of a novel autophosphorylation site (P4) on the epidermal growth factor receptor. Biochem. J. 262, 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Countaway J. L., Northwood I. C., Davis R. J. (1989) Mechanism of phosphorylation of the epidermal growth factor receptor at threonine 669. J. Biol. Chem. 264, 10828–10835 [PubMed] [Google Scholar]
- 13.Wu S. L., Kim J., Bandle R. W., Liotta L., Petricoin E., Karger B. L. (2006) Dynamic profiling of the post-translational modifications and interaction partners of epidermal growth factor receptor signaling after stimulation by epidermal growth factor using Extended Range Proteomic Analysis (ERPA). Mol. Cell. Proteomics 5, 1610–1627 [DOI] [PubMed] [Google Scholar]
- 14.Countaway J. L., Nairn A. C., Davis R. J. (1992) Mechanism of desensitization of the epidermal growth factor receptor protein-tyrosine kinase. J. Biol. Chem. 267, 1129–1140 [PubMed] [Google Scholar]
- 15.Wu W., Graves L. M., Gill G. N., Parsons S. J., Samet J. M. (2002) Src-dependent phosphorylation of the epidermal growth factor receptor on tyrosine 845 is required for zinc-induced Ras activation. J. Biol. Chem. 277, 24252–24257 [DOI] [PubMed] [Google Scholar]
- 16.Davis R. J., Czech M. P. (1985) Tumor-promoting phorbol diesters cause the phosphorylation of epidermal growth factor receptors in normal human fibroblasts at threonine-654. Proc. Natl. Acad. Sci. U.S.A. 82, 1974–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones R. B., Gordus A., Krall J. A., MacBeath G. (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 18.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 19.Huang P. H., Xu A. M., White F. M. (2009) Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2, re6. [DOI] [PubMed] [Google Scholar]
- 20.Oda K., Matsuoka Y., Funahashi A., Kitano H. (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1, 2005.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze W. X., Deng L., Mann M. (2005) Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1, 2005.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 23.Samaga R., Saez-Rodriguez J., Alexopoulos L. G., Sorger P. K., Klamt S. (2009) The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data. PLoS Comput. Biol. 5, e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu H. Y., Wittemann S., Schneider E. M., Weiss M., Joos T. O. (2008) Suspension microarrays for the identification of the response patterns in hyperinflammatory diseases. Med. Eng. Phys. 30, 976–983 [DOI] [PubMed] [Google Scholar]
- 25.Shao W., Zhou Z., Laroche I., Lu H., Zong Q., Patel D. D., Kingsmore S., Piccoli S. P. (2003) Optimization of rolling-circle amplified protein microarrays for multiplexed protein profiling. J. Biomed. Biotechnol. 2003, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]