Abstract
TIGRFAMs is a collection of protein families featuring curated multiple sequence alignments, hidden Markov models and associated information designed to support the automated functional identification of proteins by sequence homology. We introduce the term ‘equivalog’ to describe members of a set of homologous proteins that are conserved with respect to function since their last common ancestor. Related proteins are grouped into equivalog families where possible, and otherwise into protein families with other hierarchically defined homology types. TIGRFAMs currently contains over 800 protein families, available for searching or downloading at www.tigr.org/TIGRFAMs. Classification by equivalog family, where achievable, complements classification by orthology, superfamily, domain or motif. It provides the information best suited for automatic assignment of specific functions to proteins from large-scale genome sequencing projects.
INTRODUCTION
The correct assignment of protein function by homology across genomes is a difficult task. Variable evolutionary clock rates mean the most similar sequences may not be the most recently diverged. Differing patterns of gene loss across species may cause proteins of distinct function, paralogous in the last common ancestral species, to appear to be orthologous. True orthologous families may contain members with new activities. For these and other reasons, the consensus of top-scoring pairwise matches may easily misidentify a new protein. Errors in the iterated transfer of annotations among uncharacterized proteins and the relatively poor signal-to-noise ratio inherent in pairwise sequence alignment further complicate the task of protein functional identification. To address these problems, we have created a protein family resource to represent functional and not just evolutionary classifications of proteins.
Most current protein classification methods are oriented toward detection of sets of distantly related proteins that do not necessarily have the same function (1–4). In some cases, only short regions of conserved protein sequence are used to define these sets of proteins. This strategy results in inclusion of proteins that have diverse functions into a family (e.g. all proteins containing pyridoxal phosphate binding domain). A strategy that more narrowly defines families, represented in clusters of orthologous groups (COGs) (5), uses an automated clustering based on bi-directional best hit relationships across diverged species. While similar function is implied between sequences of the highest similarity of different species; conserved function is not a formal criterion used to build COGs.
BUILDING TIGRFAMs
We have built a collection of protein families, TIGRFAMs, most of which are predicted to have uniform function. The families are represented by curated multiple sequence alignments (seed alignments), hidden Markov models (HMMs) and associated annotations and cutoff scores. Models are developed using the HMMER package (6), version 2.1.1. This package allows control of HMM architecture, prior probability tables reflecting amino acid relatedness and other parameters during the building of models. Searches with the HMMs yield scores in bits that are compared to high and low stringency reference values, called the trusted and noise cutoffs. The scope of each model, that is, the set of proteins that are recognized by the HMM, is determined by which sequences are in the seed alignment, how they are aligned, the input parameters of the program hmmbuild and the cutoff value settings.
Initial clusters of related proteins from completed microbial genomes were constructed in various ways, including single linkage clustering based on all-versus-all sequence searches (7) and a BLAST (8) bi-directional best hit clustering method similar to that used in COGs (5). These initial clusters frequently contain genes of heterogeneous function. Curation of alignments, consideration of phylogenetic and/or distance trees and re-examination of protein functional assignments are performed with the objective of refining or partitioning the initial clusters into subclusters that are homogeneous in function. For each resulting subcluster, several versions of HMM are tested. Comparison of full database search results among the HMMs built from the same or different subclusters enables the selection of the best model as well as cutoff scores for each group of proteins.
Generally, a dubious member of a functionally conserved protein family is eliminated from the seed and used instead to help set an upper limit for the trusted cutoff score. Inclusion in the HMM seed would compromise the specificity of the model, since any member of the seed alignment is sure to score above any reasonable trusted cutoff score. The care taken in building these models makes them useful in predicting specific protein function and reduces the risk that incorrect historical annotations will be propagated to new sequences.
HOMOLOGY TYPES
We introduce the term ‘equivalog’ to describe proteins homologous to each other and conserved in function since their last common ancestor. Any one member of a set of orthologous proteins that differs in function from the others is not an equivalog. Sets of equivalogs, therefore, are not necessarily monophyletic. Proteins related by lateral transfer can be equivalogs, although by definition they are not orthologs.
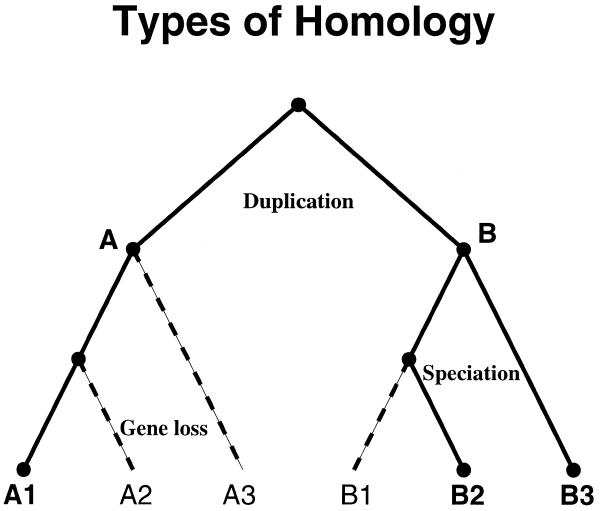
Figure 1 shows a possible phylogeny for protein evolution from a single ancestral sequence. In this model tree, paralogous proteins A and B have distinct functions. Subsequent speciation expanded each paralog into its own orthologous branch. Within each branch, the members are equivalogs if the function is conserved. The term ‘superfamily’ as introduced by Dayhoff et al. (9) and developed in the PIR-Protein Sequence Database (10) describes the complete set of proteins having sequence homology over essentially their full length. The two branches in the phylogeny belong to the same superfamily. They represent the whole of the superfamily if no other full-length homologs can be found. Otherwise, they represent a ‘subfamily’ within the superfamily. Homology may be restricted to a domain rather than the full lengths of the respective proteins. If so, the homology type is termed ‘domain’, ‘subfamily domain’ and ‘equivalog domain’, in place of superfamily, subfamily and equivalog, respectively.
Figure 1.
Homology relationships can be classified by evolutionary history, as shown in this model phylogenetic tree. The ancestral node, or root, is at the top. Duplication creates paralogs A and B with distinct function. Speciation creates an orthologous set A1, A2 and A3 from A, and B1, B2 and B3 from B. If B1, B2 and B3 share the same function, they are equivalogs as well as orthologs. Dashed lines indicate a possible pattern of gene loss that leaves only A1, B2 and B3. The resulting protein subfamily should exhibit bi-directional best hits across species but is not orthologous and does not show conserved function.
The majority of the profile HMMs in TIGRFAMs are designed to identify equivalog families from among currently available sequences. Models with other homology types have different uses. Superfamily and domain models hit relatively large numbers of proteins, provide sensitivity for the identification of remote homologs and provide insight into the possible general function of proteins whose specific role is not known. Equivalog models, in contrast, identify functionally equivalent members from larger sets of related proteins. This assignment of specific protein function is a primary goal in genome annotation.
The current TIGRFAMs dataset currently consists of 854 models, of which 516 are classified as equivalog models and 24 as equivalog domain. An additional 125 models describe small families whose function, although uncharacterized, may also be equivalent (hypothetical equivalog). The rest represent subfamily, superfamily, domain and other homology types.
EQUIVALOG HMM PERFORMANCE
Each model has a trusted cutoff, above which there should be no false positive hits, and a noise cutoff below which hits to the model are considered uninteresting. The range between trusted cutoff and noise cutoff represents scores that may or may not be true hits. Annotations attached to equivalog models for assignment to matching proteins include protein names, role categories, explanatory comments and database cross-references. Over two-thirds have been assigned prokaryotic gene symbols and nearly half have been assigned Enzyme Commission (EC) numbers. Proteins scoring above the trusted cutoff can be assigned these annotations automatically and with fairly high confidence.
The set of proteins for which equivalog models have been built is heavily weighted toward those present in published complete microbial genomes. The behavior of the equivalog model subset against genomic data suggests that these models act substantially as intended. It can be expected from first principles that most equivalog families, unlike superfamilies and domain families, will have no more than one member in most small genomes. A small genome suggests strong selective pressures against maintaining redundancies in protein function. Maintenance of distinct isozymes should be the exceptional case. Of 516 equivalog HMMs in TIGRFAMs, only 95 hit a second protein in any of the first 25 different prokaryotes whose completed genomes became available. Sixty-three of those have a second hit in exactly one genome. Only three species (Escherichia coli, Bacillus subtilis and Synechocystis sp. strain PCC 6803) have as many as three hits to any equivalog HMM. These cases generally appear to identify functionally equivalent proteins, such as the three isozymes of phospho-2-dehydro-3-deoxyheptonate aldolase found in E.coli by HMM TIGR00034.
A second test of equivalog model behavior is that the same region of the same protein should not be described by two different equivalog models. Of over 5500 predictions made by TIGRFAMs equivalog, equivalog domain and hypothetical equivalog models in 25 prokaryotic species, only B.subtilis PabB scores above the trusted cutoff for the same stretch of sequence to two different models, para-aminobenzoate synthase component I (TIGR00553) and anthranilate synthase component I, the TrpE protein of tryptophan biosynthesis (TIGR00564). Interestingly, the adjacent TrpG protein has been shown to be amphibolic, functioning in the synthesis of both tryptophan and para-aminobenzoic acid (11).
For each protein scoring above the trusted cutoff of an equivalog model, a strong prediction is made that the protein functions as described for the model. Comparison of prediction based on HMM searches to prediction based on other means (annotated protein databases, literature references and new analyses of probable protein function) is used first to pick a model from several candidates. Study may reveal functional heterogeneity among closely related proteins such that no equivalog model can be made. A subfamily or superfamily model may be made instead. After an equivalog model has been created, examination of its predictions provides feedback on model performance. TIGRFAMs has been used in annotation of microbial genomic sequences at the Institute for Genomic Research (TIGR), such as for Vibrio cholerae (12). Comparison to results from manual annotation based on multiple pairwise alignments (see www.tigr.org/CMR2/db_assignmentextver2.html for an outline of homology-based annotation standards at TIGR) has validated predictions for many models and led to improvement of a few.
USING TIGRFAMs
TIGRFAMs may be downloaded for use as a library of HMMs for protein identification or searched for text or sequence matches at its web site, http://www.tigr.org/TIGRFAMs. For any protein that scores greater than the trusted cutoff to an equivalog-type TIGRFAMs model, a prediction is made not only that this sequence shares common ancestry with the members of the seed alignment, but also that it shares a common function. Pre-calculated results of HMM searches with TIGRFAMs models against a collection of completed microbial genomes can be found in the Comprehensive Microbial Resource (CMR) (13) accessible from the CMR homepage www.tigr.org/CMR. The assigned homology type (equivalog or other), associated annotations, seed alignments, full alignments and tables of hits to microbial genomic sequences are presented for each model.
The TIGRFAMs collection is intended to complement the Pfam A collection (1) of profile HMMs. It uses the same scoring system, the same suite of programs for generating and using HMMs and a similar representation for ancillary data. The two sets of models may be combined in a single library and searched simultaneously.
Among Pfam HMMs with at least one trusted hit to protein sequence from a complete prokaryotic genome, the models hit on average just over 20 hits in 25 genomes. Among TIGRFAMs equivalog, equivalog domain and hypothetical equivalog HMMs, the average is just under 10 hits in 25 genomes. For proteins hit by both, the TIGRFAMs equivalog model usually describes a family equal in size to or smaller than the overlapping Pfam model, and only rarely larger. The TIGRFAMs model hit region averages ∼40% longer than the corresponding Pfam hit region and is only rarely shorter. Longer hits to fewer proteins are expected for models designed for functional identification of whole proteins rather than detection of domain and superfamily relationships.
Several TIGRFAMs equivalog models may describe different branches of a superfamily or domain family described by a single Pfam model. Other proteins from the same superfamily may fail to score above the cutoff value for any current equivalog model. This amounts to a negative prediction, a warning that calling these orphan proteins functionally equivalent to those within the scope of an equivalog model may be unwarranted.
Using the domain and superfamily classification systems of Pfam HMMs, PIR, COGs and other resources for the general classification of proteins has undeniable value in support of the prediction of protein function by homology. Addition of the specific functional predictions afforded by TIGRFAMs equivalog HMMs offers a hierarchical classification system that should enhance both the automation and the accuracy of protein annotation.
References
- 1.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam Protein Families Database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasarao G.Y., Yeh,L.S., Marzec,C.R., Orcutt,B.C. and Barker,W.C. (1999) PIR-ALN: a database of protein sequence alignments. Bioinformatics, 15, 382–390. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. Updated article in this issue: Nucleic Acids Res. (2001), 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy S.R.(1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 7.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaffer,A.A, Zhang,J., Zhang. Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dayhoff M.O. (1976) The origin and evolution of protein superfamilies. Fed. Proc., 35, 2132–2138. [PubMed] [Google Scholar]
- 10.Barker W.C., Pfeiffer,F. and George,D.G. (1996) Superfamily classification in PIR-International Protein Sequence Database. Methods Enzymol., 266, 59–71. [DOI] [PubMed] [Google Scholar]
- 11.Slock J., Stahly,D.P., Han,C.Y., Six,E.W. and Crawford,I.P. (1990) An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J. Bacteriol., 172, 7211–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidelberg J.F., Eisen,J.A., Nelson,W.C., Clayton,R.A., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Umayam,L., Gill,S.R., Nelson,K.E., Read,T.D., Tettelin,H., Richardson,D., Ermolaeva,M.D., Vamathevan,J., Bass,S., Qin,H., Dragoi,I., Sellers,P., McDonald,L., Utterback,T., Fleishmann,R.D., Nierman,W.C. and White,O. (2000) DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature, 406, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson J.D., Umayam,L.A., Hickey,E.K. and White,O. (2001) The Comprehensive Microbial Resource. Nucleic Acids Res., 29, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]