Abstract
A new thermodynamic database for normal and modified nucleic acids has been developed. This Thermodynamic Database for Nucleic Acids (NTDB) includes sequence, structure and thermodynamic information as well as experimental methods and conditions. In this release, there are 1851 sequences containing both normal and modified nucleic acids. A user-friendly web-based interface has been developed to allow data searching under different conditions. Useful thermodynamic tools for the study of nucleic acids have been collected and linked for easy usage. NTDB is available at http://ntdb.chem.cuhk.edu.hk.
INTRODUCTION
Thermodynamic properties of nucleic acids have attracted considerable attention since the early 1960s. Information on thermodynamics is very important in understanding the biological function of nucleic acids, such as DNA replication, mutation, repair and transcription (1,2). Its availability is also essential for the design of optimal experimental conditions for carrying out numerous molecular biological techniques including antigene targeting (3–5), Kunkel mutagenesis (6), mismatch amplification refractory mutation assay (7), oligonucleotide chip design (8–11), PCR (12), sequencing by hybridization (8,13) and Southern blotting (14). It is also helpful in the development of secondary structure prediction of nucleic acids (15,16), and plays a significant role in the design of novel oligonucleotides modification for antisense therapeutics (17).
With the rapid development of Internet technology, the availability of a compiled source of thermodynamics information in the form of a database would greatly enhance scientific communication and the sharing of scientific results. However, most web-accessible biological databases are focused on sequences and structures. Until recently, Gromiha et al. developed a Thermodynamic Database for Proteins and Mutants (ProTherm), emphasizing collection of thermodynamic and structural information on protein mutant stability (18). There is no similar web-accessible database whereby the thermodynamic information of normal and modified nucleic acids has been categorically collected. In this work, we present a Thermodynamic Database for Nucleic Acids (NTDB), which includes a collection of thermodynamic and structural data, experimental methods, conditions and the corresponding references.
DATABASE INFRASTRUCTURE
In NTDB, each entry is given an identity number. The information in each entry includes: (i) the sequence of normal and modified nucleic acids; (ii) the structural type (e.g. DNA or RNA), with or without mismatches, dangling ends, bulge loops and modification information on the base, sugar and/or phosphate position; (iii) thermodynamic data including the enthalpy, ΔH; entropy, ΔS; Gibbs free energy, ΔG, and also melting temperature, Tm; (iv) the type of measuring instrument, strand concentration, composition of the buffer used (e.g. NaCl, sodium cacodylate, sodium phosphate, EDTA); and (v) relevant reference information. An example is shown in Figure 1. The reference for all entries in NTDB is linked to the NCBI PubMed literature database (http://www.ncbi.nlm.nih.gov/Entrez/medline.html). In addition, a set of useful tools such as thermal denaturation profile, extinction coefficient, free energy calculation, secondary structure prediction and primer selection relevant to the study of nucleic acids are also collected for the user. Links are implemented to connect some valuable related articles as well as information on scientific groups active in this field.
Figure 1.
An example of an entry in NTDB.
DATABASE STATISTICS
In this release, NTDB contains 1851 entries, which are classified into 49 different classes. It includes 727 DNA, 162 RNA, 79 DNA/RNA, 37 PNA and 105 other modified nucleic acid sequences. There are 287 entries with base modifications and 147 entries with other modifications among the 1851 entries. In addition, there are 532 entries containing mismatches, 373 entries with dangling ends, 58 entries with bulge loops, 207 hairpin entries and 47 entries on DNA and RNA triplexes.
NTDB SEARCHING
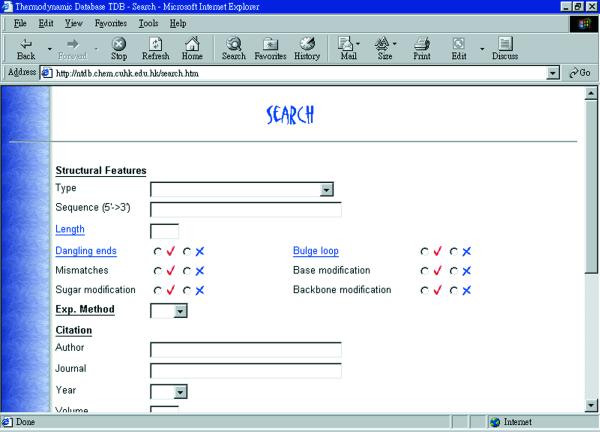
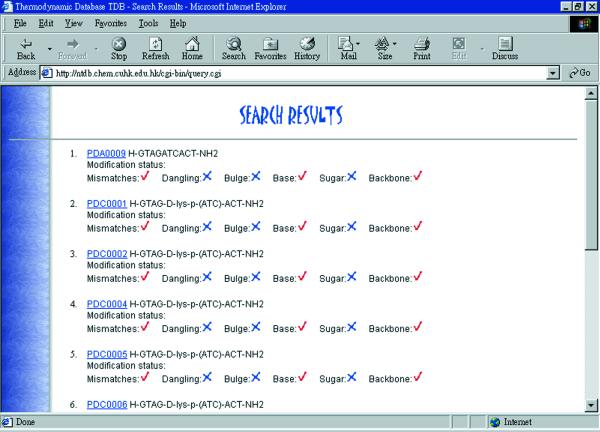
A web-based interface has been developed for searching information in the NTDB as shown in Figure 2. A search can be readily carried out when appropriate criteria are imposed, e.g. structural type, sequence, mismatches, dangling and bulge loop information, other modification information, range of ΔH, ΔS, Tm, citation information. For each search, a shortlist of sequences fulfilling the search requirements will be shown (Fig. 3) and the information is linked to the NTDB ID.
Figure 2.
The searching interface of NTDB.
Figure 3.
An example of a search result using NTDB.
NTDB AVAILABILITY AND CITATION
NTDB can be freely accessed at http://ntdb.chem.cuhk.edu.hk. NTDB users are encouraged to cite this article and include the URL address. Any comments, suggestions and contributions are welcomed and should be addressed to the corresponding author.
References
- 1.Mendelman L.V., Boosalis,M.S., Petruska,J. and Goodman,M.F. (1989) Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem., 264, 14415–14423. [PubMed] [Google Scholar]
- 2.Petruska J., Goodman,M.F., Boosalis,M.S., Sowers,L.S., Cheong,C. and Tinoco,I.,Jr (1988) Comparison of DNA Melting Thermodynamics and DNA Polymerase Fidelity. Proc. Natl Acad. Sci. USA, 85, 6252–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frier S.M. (1993) Hybridization: Considerations Affecting Antisense Drugs. In Crooke,S.T. and Lebleu,B. (eds), Antisense Research and Applications. CRC Press, Boca Raton, FL, pp 67–82.
- 4.Zon G. (1989) Pharmaceutical considerations. In Cohen,J.S. (ed.), Oligonucleotides: antisense inhibitors of gene expression. CRC Press, Boca Raton, FL, pp 233–249.
- 5.Symons R.H. (1989) Nucleic Acids Probes. CRC Press, Boca Raton, FL.
- 6.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 7.Cebula T.A., Payne,W.L. and Feng,P. (1995) Simultaneous Identification of Strains of Escherichia coli Serotype O157:H7 and Their Shiga-Like Toxin Type by Mismatch Amplification Mutation Assay-Multiplex PCR. J. Clin. Microbiol., 33, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor S.P.A., Rava,R.P., Huang,X.C., Pease,A.C., Holmes,C.P. and Adams,C.L. (1993) Multiplexed Biochemical Assays with Biological Chips. Nature, 364, 555–556. [DOI] [PubMed] [Google Scholar]
- 9.Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Accessing genetic information with high-density DNA arrays. Science, 274, 610–614. [DOI] [PubMed] [Google Scholar]
- 10.Pease A.C., Solas,D., Sullivan,E.J., Cronin,M.T., Holmes,C.P. and Fodor,S.P.A. (1994) Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl Acad. Sci. USA, 91, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapolsky R.J. and Lipshutz,R.J. (1996) Mapping genomic library clones using oligonucleotide arrays. Genomics, 33, 445–456. [DOI] [PubMed] [Google Scholar]
- 12.Saiki R.K., Gelfand,D.H., Stoffel,S., Scharf,S., Higuchi,R.H., Horn,G.T., Mullis,K.B. and Erlich,H.A. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–494. [DOI] [PubMed] [Google Scholar]
- 13.Parinov S., Barsky,V., Yershov,G., Kirillov,E., Timofeev,E., Belgovskiy,A. and Mirzabekov,A. (1996) DNA sequencing by hybridization to microchip octa- and decanucleotides extended by stacked pentanucleotides. Nucleic Acids Res., 24, 2998–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Southern E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol., 98, 503–517. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 16.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 17.Freier S.M. and Altmann K.-H. (1997) The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res., 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromiha M.M., An,J., Kono,H., Oobatake,M., Uedaira,H., Prabakaran,P. and Sarai A. (2000) ProTherm, version 2.0: Thermodynamic database for proteins and mutants. Nucleic Acids Res., 28, 283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]