Abstract
Hepatocyte nuclear factor 4 alpha (HNF4a) is a liver-enriched master regulator of liver function. HNF4a is important in regulating hepatic expression of certain cytochrome P450s. The purpose of this study was to use mice lacking HNF4a expression in liver (HNF4a-HNull) to elucidate the role of HNF4a in regulating hepatic expression of phase II enzymes and transporters in mice. Compared with male wild-type mice, HNF4a-HNull male mouse livers had (1) markedly lower messenger RNAs (mRNAs) encoding the uptake transporters sodium taurocholate cotransporting polypeptide, organic anion transporting polypeptide (Oatp) 1a1, Oatp2b1, organic anion transporter 2, sodium phosphate cotransporter type 1, sulfate anion transporter 1, sodium-dependent vitamin C transporter 1, the phase II enzymes Uridine 5′-diphospho (UDP)-glucuronosyltransferase (Ugt) 2a3, Ugt2b1, Ugt3a1, Ugt3a2, sulfotransferase (Sult) 1a1, Sult1b1, Sult5a1, the efflux transporters multidrug resistance–associated protein (Mrp) 6, and multidrug and toxin extrusion 1; (2) moderately lower mRNAs encoding Oatp1b2, organic cation transporter (Oct) 1, Ugt1a5, Ugt1a9, glutathione S-transferase (Gst) m4, Gstm6, and breast cancer resistance protein; but (3) higher mRNAs encoding Oatp1a4, Octn2, Ugt1a1, Sult1e1, Sult2a2, Gsta4, Gstm1-m3, multidrug resistance protein (Mdr) 1a, Mrp3, and Mrp4. Hepatic signaling of nuclear factor E2–related factor 2 and pregnane X receptor appear to be activated in HNF4a-HNull mice. In conclusion, HNF4a deficiency markedly alters hepatic mRNA expression of a large number of phase II enzymes and transporters, probably because of the loss of HNF4a, which is a transactivator and a determinant of gender-specific expression and/or adaptive activation of signaling pathways important in hepatic regulation of these phase II enzymes and transporters.
Keywords: HNF4a, knockout, Ugt, Gst, Sult, transporter
Hepatocyte nuclear factor 4 alpha (HNF4a) is a liver-enriched master regulator of liver development and differentiation. HNF4a is essential for hepatocyte differentiation in fetal liver (Kyrmizi et al., 2006; Li et al., 2000) and maintenance of liver function in adult (Gonzalez, 2008; Hayhurst et al., 2001). HNF4a directly binds to a large number of gene promoters in human and mouse liver (Odom et al., 2004, 2007; Schmidt et al., 2010). Mice with hepatocyte-specific null (HNull) of HNF4a have fatty liver and markedly increased unconjugated bile acids and ammonia, thus revealing a critical role of HNF4a in regulating hepatic metabolism of fatty acids, bile acids, and ureagenesis (Hayhurst et al., 2001; Inoue et al., 2002, 2004, 2006).
Humans have very large individual variations in hepatic basal expression of HNF4a (Wortham et al., 2007), and mutation of HNF4a causes maturity-onset diabetes of the young humans (MODY1) (Ryffel, 2001). HNF4a is markedly downregulated by hypoxia in HepG2 hepatoma cells (Mazure et al., 2001). Hepatic expression and/or the transcriptional activity of HNF4a is decreased markedly in severe cirrhotic human livers, alcoholic liver disease, tumor necrosis factor-α–induced hepatotoxicity, and hepatoma progression (Berasain et al., 2003; Kang et al., 2009; Lazarevich et al., 2004; Zhou et al., 2007). Therefore, it is important to understand how deficiency of HNF4a influences hepatic gene expression and its underlying mechanism.
Previous studies indicate that HNF4a has a critical role in regulating hepatic expression of drug-processing genes, namely cytochrome P450s (P450s/Cyps), phase II conjugation enzymes, and transporters. Knockdown of HNF4a in primary human hepatocytes using small interfering RNA (siRNA) results in decreased messenger RNA (mRNA) levels of various P450s, Uridine 5′-diphospho (UDP)-glucuronosyltransferase (UGT) 1A1, UGT1A9, sulfotransferase (SULT) 2A1, multidrug resistance protein (MDR) 1, bile salt export pump (BSEP), multidrug resistance–associated protein (MRP) 2, organic anion transporting polypeptide (OATP) 1B1, and organic cation transporter 1 (OCT1), as well as xenobiotic receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) (Kamiyama et al., 2007). Hepatic mRNA expression of diverse drug metabolizing enzymes and transporters correlates with the expression of HNF4a in human livers (Wortham et al., 2007). HNF4a-HNull mice have dramatic downregulation of male-specific and female-specific P450s, such as Cyp2d9 in males and Cyp2b10, Cyp2b13, Cyp3a41, and Cyp3a44 in females (Wiwi et al., 2004). Interestingly, the female-specific Cyp2b9 was markedly induced in male HNF4a-HNull mice. In contrast, certain P450s that display minimal gender-divergent expression, such as Cyp3a11 and 3a25, were moderately decreased in both genders of HNF4a-HNull mice (Wiwi et al., 2004). HNF4a is proposed to contribute to hepatic gender-specific gene expression by positively regulating a subset of male-specific genes, whereas concomitantly inhibiting the expression of certain female-specific genes (Wiwi et al., 2004).
Despite the information about the regulation of hepatic P450s by HNF4a (Holloway et al., 2006; Jover et al., 2009; Wiwi et al., 2004), little is known regarding the role of HNF4a in regulating hepatic expression of phase II conjugation enzymes and xenobiotic transporters in mice. Thus, the purpose of this study was to use HNF4a-HNull mice to elucidate the role of HNF4a in the regulation of hepatic expression of major phase II conjugation enzymes and xenobiotic transporters, namely Ugts, Sults, glutathione S-transferases (Gsts), as well as uptake and efflux transporters.
MATERIALS AND METHODS
HNF4a-HNull mice.
Mice were maintained at an American Animal Associations Laboratory Animal Care-accredited facility at the University of Kansas Medical Center. HNF4a-HNull mice were generated with an albumin promoter–regulated Cre (Alb-Cre)-loxP-mediated deletion of exons 4 and 5 of the HNF4a gene (Hayhurst et al., 2001). Age-matched young-adult HNF4a-HNull mice (HNF4a flox/flox, Alb-Cre/+) and their wild-type (WT) control littermates (HNF4a flox/flox, Alb-Cre/−) were fed rodent chow (#8064, Teklad; Harlan, Indianapolis, IN). Mice were housed at an ambient temperature of 22°C with alternating 12-h light/dark cycles and allowed water and feed ad libitum. In our previous studies on ontogeny of hepatic and renal mRNA expression of drug-processing genes, male and female mice of 45 days of age were used. Additionally, male HNF4a-Hnull mice of 45 days of age were used in a previous study (Hayhurst et al., 2001). Moreover, HNF4a-Hnull mice have mortality > 70% by 8 weeks of age (Hayhurst et al., 2001). Thus, liver tissues from 45-day-old WT control and HNF4a-HNull male and female mice (n = 5–6 for each group) were used in this study. Liver and kidney tissues were snap frozen in liquid nitrogen at the time of collection and stored at −80°C until use. All animal procedures in this study were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center.
Determination of mRNA expression with quantigene plex assay.
Total RNA of liver and kidney was isolated using RNA-Bee reagent (Tel-Test, Inc., Friendswood, TX). Mouse liver mRNAs encoding phase II enzymes (Ugts, Sults, and Gsts) and transporters was determined from total RNA of livers as described by the manufacturer's protocol utilizing Quantigene Plex 1.0 Technology (Panomics/Affymetrix, Fremont, CA). Individual bead-based oligonucleotide probe sets, specific for each gene examined, were developed by Panomics/Affymetrix (Plex 2055 for Ugts/Gsts, Plex 2061 for Sults, and Plex 2046 for transporters). Genes and accession numbers are available at http://www.panomics.com. Samples were analyzed using a Bio-Plex 200 System Array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex Data Manager Software Version 5.0 (Bio-Rad). All data were standardized to the internal control, glyceraldehyde 3-phosphate dehydrogenase. Ugt, Sult, and Gst mRNAs were reported as relative mRNA expression with values of male WT controls set at 1.0.
Determination of expression of individual mRNAs with branched DNA signal amplification assay.
The mRNA of individual genes was quantified using Quantigene branched DNA (bDNA) signal amplification kit (Panomics/Affymetrix, Fremont, CA) with modifications (Hartley and Klaassen, 2000). The sequences of the bDNA probes for most of these drug-processing and nuclear receptor genes have been reported previously (Alnouti and Klaassen, 2006; Alnouti et al., 2006; Buist and Klaassen, 2004; Cheng and Klaassen, 2006; Cheng et al., 2005, 2007; Cui et al., 2009; Maher et al., 2005; Petrick and Klaassen, 2007), with the exception of sulfate anion transporter 1 (Sat1) and sodium-dependent vitamin C transporter (Svct) 1, which are provided in the Supplementary table 2. Luminescence of samples in 96-well plates was determined with a Synergy 2 Microplate reader (BioTek Instruments, Inc., Winooski, VT) and reported as relative mRNA expression with values of male WT controls set at 1.0.
Determination of DNA binding of transcription factors with procarta TF plex assays.
The in vitro binding of transcription factors (TFs), namely peroxisome proliferator–activated receptor (PPAR), PXR, glucocorticoid receptor (GR)/progestone receptor (PR), HNF1, CCAAT/enhancer binding protein (C/EBP), and nuclear factor κB (NF-κB) to their consensus cis-response elements was determined with “By Request” Procarta fluorescent microsphere-based TF plex assays (Yaoi et al., 2006) (catalog # PC5903 and PC5904; Panomics/Affymetrix) using liver nuclear extracts prepared from male HNF4a-Hnull and WT mice. The PPAR assays determine the binding of TFs to the consensus PPAR cis-response elements and thus do not differentiate the DNA binding of PPARα from other PPAR family members, including PPARβ and PPARγ. Similarly, the C/EBP assays determine the binding of TFs to the consensus cis-response elements of C/EBP family members, including C/EBPα, C/EBPβ, and C/EBPγ. All TF plex kits contain all the reagents required to detect the presence of activated TFs for analysis on the Luminex system. Samples were analyzed using a Bio-Plex 200 System Array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex Data Manager Software Version 5.0 (Bio-Rad).
Statistics.
Data are presented as mean ± SE. Differences between various groups were determined using ANOVA followed by post hoc test, with significance set at p < 0.05.
RESULTS
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Uptake Transporters
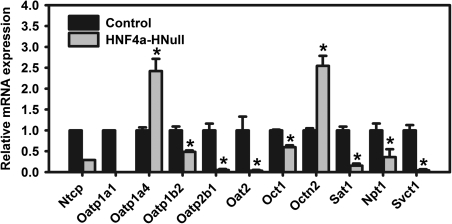
The loss of HNF4a had marked effects on hepatic expression of uptake transporters. Of the 11 uptake transporters examined, 9 were markedly decreased and 2 more than doubled (Fig. 1). Consistent with a previous report (Hayhurst et al., 2001), hepatic sodium taurocholate cotransporting polypeptide (Ntcp) and Oatp1a1 were markedly lower in HNF4a-HNull male mice (Fig. 1). Hepatic Ntcp and Oatp1a1 mRNAs were also much lower in female HNF4a-HNull mice (data not shown). In contrast, Oatp1a4 mRNA was 142% higher in male (Fig. 1) but tended to be lower in female (data not shown) HNF4a-HNull mice, resulting in a loss of gender-divergent expression of Oatp1a4. Hepatic Oatp1b2 and Oatp2b1 mRNAs were 51 and 94% lower in male HNF4a-HNull mice, respectively. Hepatic expression of Oat2, the only Oat family member with appreciable expression in mouse liver (Buist and Klaassen, 2004), was much lower in male HNF4a-HNull mice than male WT mice. Hepatic Oct1 mRNA was 40% lower, whereas Octn2 mRNA was 154% higher in male HNF4a-HNull mice.
FIG. 1.
Hepatic mRNAs encoding uptake transporters in adult male mice with liver-specific deletion (HNull) of HNF4a. The y-axis represents relative mRNA expression with values of WT controls set at 1.0. Data without error bar represent transporter mRNAs in pooled total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) determined by the Quantigene Plex assay. Data with error bar represent transporter mRNAs in individual samples of total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) determined by the bDNA assay. Mean ± SE. *p < 0.05 compared with WT control.
Sat1/Slc26a1 is a liver-enriched sinusoidal transporter responsible for the uptake of sulfate into hepatocytes (Markovich and Aronson, 2007). The sodium-dependent phosphate cotransporter 1 (Npt1/Slc17a1) is important in hepatic uptake of phosphate. Hepatic Sat1 and Npt1 mRNAs were 85 and 64% lower, respectively, in male HNF4a-HNull mice than male WT mice.
There are two isoforms of ascorbic acid transporters, the low-affinity high-capacity transporter SVCT1 (Slc23a1) and the high-affinity low-capacity transporter SVCT2 (Slc23a2) (Savini et al., 2008). Svct2 is essential for vitamin C transport into brain and perinatal survival in mice (Sotiriou et al., 2002). Svct1 is expressed at much higher levels than Svct2 in liver and kidney and is considered as the major ascorbic acid transporter in liver (Kuo et al., 2004). Hepatic Svct1 mRNA was markedly downregulated (96%) in HNF4a-HNull mice.
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Ugts
The only Ugt that was more highly expressed in HNF4a-HNull than WT mice was Ugt1a1 by 79% (Fig. 2). Seven of the 10 Ugts had a lower expression in HNF4a-HNull mice: Ugt1a5 55%, Ugt1a9 64%, Ugt2a3 84%, Ugt2b1 94%, Ugt2b36 44%, Ugt3a1 86%, and Ugt3a2 96% lower than WT mice. In contrast, HNF4a-HNull mice had unaltered hepatic mRNAs encoding Ugt1a6, Ugt2b34, Ugt2b35, as well as UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase (Ugp2), which catalyzes the formation of UDP-glucuronic acid, the cosubstrate for glucuronidation.
FIG. 2.
Hepatic mRNAs encoding Ugts in adult male mice with liver-specific deletion (HNull) of HNF4a. Ugt mRNAs in total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) were determined by the Quantigene Plex assay. The y-axis represents relative mRNA expression of Ugts with values of WT controls set at 1.0. Mean ± SE. *p < 0.05 compared with WT control.
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Sults
HNF4a-HNull male mice had 69, 95, and 73% lower Sult1a1, Sult1b1, and Sult5a1 mRNA but 30- and 111-fold higher Sult1e1 and Sult2a2 mRNA, respectively, than WT mice (Fig. 3). Hepatic mRNA expression of Sult1c1 and Sult1d1 remained unchanged in HNF4a-HNull mice. The sulfate donor synthesis enzyme 3′-phosphoadenosine 5′-phosphosulfate synthetase (Papss) has two isoforms, Papss1 and Papss2; Papss2 is the predominant isoform in mouse liver (Alnouti and Klaassen, 2006). Hepatic Papss2 mRNA remained unchanged, whereas Papss1 mRNA was 48% higher in HNF4a-HNull mice (Fig. 3).
FIG. 3.
Hepatic mRNAs encoding Sults in adult male mice with liver-specific deletion (HNull) of HNF4a. Sult mRNAs in total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) were determined by the Quantigene Plex assay. The y-axis represents relative mRNA expression of Sults with values of WT controls set at 1.0. Mean ± SE. *p < 0.05 compared with WT control.
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Gsts
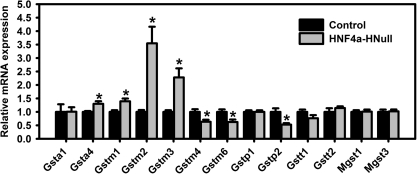
In general, the lack of HNF4a had less of an effect on Gsts than on Ugts and Sults. The most marked changes were a 255% increase of Gstm2 and 128% increase in Gstm3 in the HNF4a-HNull male mice (Fig. 4). HNF4a deficiency also increased Gsta4 30% and Gstm1 39%, whereas it decreased Gstm4 37%, Gstm6 38%, and Gstp2 47% (Fig. 4). In contrast, HNF4a had no effect on the other half of the Gsts, namely Gsta1, Gstp1, Gstt1, Gstt2, as well as microsomal Gst1 (mGst1) and mGst3 (Fig. 4).
FIG. 4.
Hepatic mRNAs encoding Gsts in adult male mice with liver-specific deletion (HNull) of HNF4a. Gst mRNAs in total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) were determined by the Quantigene Plex assay. The y-axis represents relative mRNA expression of Gsts with values of WT controls set at 1.0. Mean ± SE. *p < 0.05 compared with WT control.
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Efflux Transporters
Of the 15 hepatic efflux transporters examined, the absence of HNF4a increased the expression of 3 and decreased the expression of 3 transporters. Hepatic Mrp3 and Mrp4 mRNAs were 249 and 179% higher, respectively, whereas Mrp6 mRNA was 66% lower in HNF4a-HNull than WT male mice (Fig. 5). Male HNF4a-HNull mice had 221% higher Mdr1a mRNA but 64% lower breast cancer resistance protein (Bcrp, Abcg2) mRNA. Remarkably, hepatic multidrug and toxin extrusion 1 (Mate1/Slc47a1) mRNA was 96% lower in male HNF4a-HNull mice. In contrast, hepatic mRNA expression of Mrp2, Mdr2, Bsep, Abcg5, Abcg8, Mate2, organic solute transporter α (Ostα), Abca1, and Atp8b1 remained largely unaltered by HNF4a deficiency (Fig. 5).
FIG. 5.
Hepatic mRNAs encoding efflux transporters in adult male mice with liver-specific deletion (HNull) of HNF4a. The y-axis represents relative mRNA expression with values of WT controls set at 1.0. Data without error bar represent transporter mRNAs in pooled total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) determined by the Quantigene Plex assay. Data with error bar represent transporter mRNAs in individual samples of total RNA from livers of HNF4a-HNull and WT control mice (N = 5–6) determined by the bDNA assay. Mean ± SE. *p < 0.05 compared with WT control.
Effects of HNF4a Deficiency in Liver on Gender-Divergent Renal Transporters
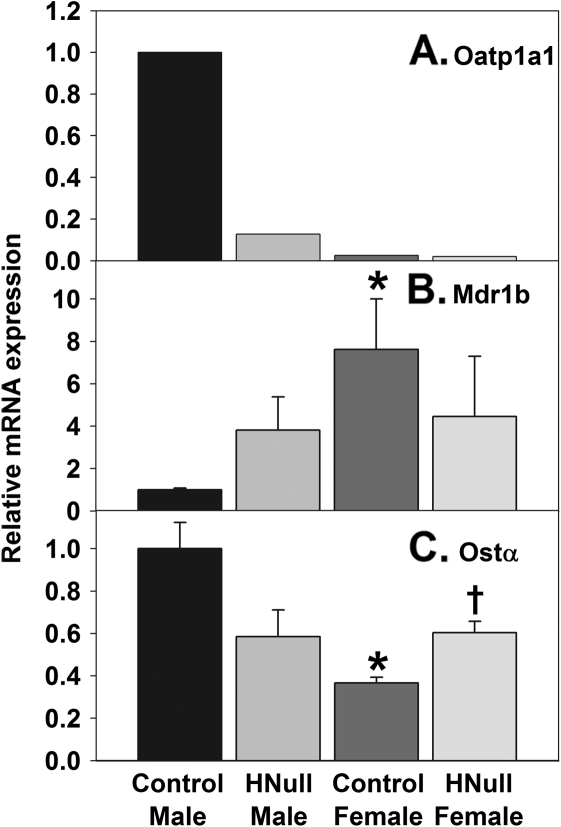
Because many gender-specific patterns of gene expression were lost in livers of HNF4a-HNull mice (Holloway et al., 2008), renal expression of a few gender-divergent genes were quantified to determine whether HNF4a deficiency in liver affects renal gender-divergent gene expression. Oatp1a1 is male predominant in mouse kidney (Cheng et al., 2005). Renal Oatp1a1 mRNA was downregulated markedly in male HNF4a-HNull mice (Fig. 6A). Compared with corresponding WT mice, renal Mdr1b mRNA tended to be higher (282%) in male HNF4a-HNull mice but lower (42%) in female HNF4a-HNull mice, resulting in a loss of gender-divergent expression of Mdr1b in HNF4a-HNull mice (Fig. 6B). Renal expression of Ostα was 173% higher in male than female WT mice (Fig. 6C). Ostα mRNA in kidneys tended to be lower (42%, p = 0.07) in HNF4a-HNull male mice than in WT males, but Ostα was significantly higher (65%) in HNF4a-HNull female mice than in WT females, resulting in a loss of gender-divergent expression of Ostα in HNF4a-HNull mice (Fig. 6C). In contrast, HNF4a-HNull mice had unaltered renal mRNAs encoding xenobiotic transporters that lack gender-divergent expression (Alnouti et al., 2006; Maher et al., 2005; Tanaka et al., 2005), such as Oct1, Octn2, Mrp1, Mrp2, Mrp5, Bcrp, and Mate1 (data not shown).
FIG. 6.
Renal mRNAs encoding transporters in adult male and female mice with liver-specific deletion (HNull) of HNF4a. The y-axis represents relative mRNA expression with values of male WT controls set at 1.0. Data with error bar represent transporter mRNAs in individual samples of total RNA, whereas data without error bar represent transporter mRNAs in pooled total RNA, from kidneys of HNF4a-HNull and WT control mice (N = 5–6) determined by the bDNA assay. Mean ± SE. *p < 0.05 compared with WT male mice; †p < 0.05 compared with WT female mice.
Effects of HNF4a Deficiency on Hepatic mRNAs Encoding Nuclear Receptors and Other Transcription Factors Important in the Regulation of Drug-Processing Genes
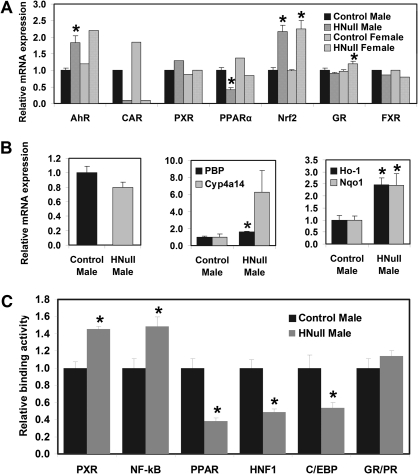
Very similar changes in hepatic mRNA expression of nuclear receptors and TFs were observed in male and female HNF4a-HNull mice (Fig. 7A). Hepatic aryl hydrocarbon receptor (AhR) mRNA was about 75% higher in HNF4a-HNull than WT mice. HNF4a deficiency in the liver almost totally abolished the expression of CAR, decreased PPARα about 55%, whereas expression of mRNAs encoding PXR, GR, and farnesoid X receptor (FXR) remained largely unchanged in HNF4a-HNull mice (Fig. 7A). Nuclear factor E2–related factor 2 (Nrf2) mRNA was about 120% higher in HNF4a-HNull than WT mice (Fig. 7A). In contrast, hepatic HNF1a mRNA tended to be lower in male HNF4a-HNull mice (Fig. 7B, left).
FIG. 7.
(A–B) Hepatic mRNA expression of nuclear receptors/TFs important in the regulation of drug-processing genes in adult male and/or female mice with liver-specific deletion (HNull) of HNF4a. The y-axis represents relative mRNA expression with values of male WT controls set at 1.0. Data with error bar represent mRNAs in individual samples of total RNA, whereas data without error bar represent mRNAs in pooled total RNA, from livers of HNF4a-HNull and WT control mice (N = 5–6) determined by the bDNA assay. (C) Binding of TFs to their consensus cis-response elements. Nuclear extracts from livers of male HNF4a-Hnull and WT mice (N = 5) were used to determine in vitro DNA binding of TFs with Procarta TF binding assays. Mean ± SE. *p < 0.05 compared with WT control.
Although PPARα mRNA was lower in male HNF4a-HNull mice, the transcription coactivator of PPARα, PPAR-binding protein (PBP) mRNA was 58% higher, and the PPARα target gene Cyp4a14 mRNA was much higher than that in WT mice (Fig. 7B, middle).
Consistent with induction of Nrf2 mRNA, hepatic mRNA expression of two Nrf2 target genes, heme oxygenase-1 (Ho-1) and NAD(P)H quinone oxidoreductase 1 (Nqo1), were ∼2.4-fold higher in male HNF4a-HNull mice than their WT controls (Fig. 7B, right), indicating that hepatic Nrf2 signaling is enhanced by HNF4a deficiency.
Effects of HNF4a Deficiency on Activities of Binding of TFs to Their Cis-Response Elements
The Procarta TF assays were used to determine the activation of TFs by quantifying their in vitro binding to their consensus cis-response elements. DNA binding of PXR and NF-κB in nuclear extracts from male HNF4a-null livers was 46 and 49% higher, whereas binding of PPAR (including all three PPAR family members, namely PPARα, PPARβ, and PPARγ), HNF1, and C/EBP (including all three C/EBP family members, namely C/EBPα, C/EBPβ, and C/EBPγ) to their consensus response elements in male HNF4a-null livers was 62, 51, and 46% lower, respectively, than male WT mice. DNA binding of GR/PR remained unchanged in male HNF4a-null livers (Fig. 7C).
DISCUSSION
The present data indicate that HNF4a has a critical role in regulating hepatic expression of certain Ugts and Sults as well as many important uptake and efflux transporters responsible for the hepatic disposition of xenobiotics. Moreover, loss of HNF4a in liver causes adaptive activation of certain nuclear receptors in liver as well as alterations in systemic hormonal signaling, resulting in selective loss of gender-divergent renal expression of certain uptake and efflux transporters.
HNF4a regulates gene expression by binding to direct repeat (DR) motifs of the RG(G/T)TCA sequence separated by one nucleotide (DR1) (Nishiyama et al., 1998). However, a recent study of genome-wide prediction of HNF4a functional binding sites demonstrates that in addition to DR1, HNF4a also binds to DR2 and other sites (Kel et al., 2008). In this study, we used NHRSCAN (Sandelin and Wasserman, 2005), a web-based program for the prediction of nuclear receptor–binding sites to search for DR1 and DR2 sites as potential DNA-binding sites of HNF4a. As shown in Table 1, the majority of the enzymes and transporters markedly downregulated by HNF4a deficiency have DR1 sites identified either within 2 kb of the proximal promoter or in the first intron of these genes, suggesting that HNF4a may directly bind to DR1/DR2 sites to activate hepatic expression of these genes. In contrast, genes markedly increased by HNF4a deficiency, such as Sult1e1 and Sult2a2, do not have DR1 sites in either the proximal promoter or the first intron of these genes, suggesting that the marked induction of these genes by HNF4a deficiency may be because of an indirect mechanism, such as removal of suppressors or activation of transactivators for these genes.
TABLE 1.
Bioinformatic Analysis of the DR1/DR2 Sites in the Proximal Promoters (2 kb) and Intron1-2 of Drug-Processing Genes That Are Markedly Downregulated in Livers of HNF4a-HNull Mice
| Gene name | DR1/DR2 site sequence | DR1/DR2 site location |
| Ugt2a3 | DR1 TGAACTCTGCTCT | Intron1-2 |
| DR2 TGGCCTAGTCACCC | −346 bp | |
| Ugt2b1 | DR1 TGAACCTTGAGCT | −144 bp |
| Ugt2b36 | DR1 TTGTCAAAGGTCA, | −1978 bp |
| DR1 AGATAAAAGGTCA | −31 bp | |
| Ugt3a1 | DR1 AGGTAATAGGTCA, | Intron1-2 |
| DR1 TGACCTCTCAGCT, | Intron1-2 | |
| DR2 AGGGCAAAAGGGCA | −34 bp | |
| Ugt3a2 | DR1 GAAACTTTGCAC | −855 bp |
| Sult1a1 | DR1 TGAACTCAGACCA | −1717 bp |
| DR1 TGTCCTATAACCC | −1024 bp | |
| DR2 AGGTCAAGGGGGCA | −592 bp | |
| Sult1b1 | DR1 TGAACTTTAAGCC | Intron1-2 |
| DR1 TGTCCTCTGTCCT | Intron1-2 | |
| Gstm6 | DR1 TGGCCCTTCACCT | −1990 bp |
| Oat2 | DR1 AGCTCACAGGCCA | −1521 bp |
| DR1 AGGCCAGGGTTCA | −1240 bp | |
| Oatp1a1 | DR1 TGACCTATGATCT | −300 bp |
| Oatp2b1 | DR1 GGATCAAAGGCCA | Intron1-2 |
| DR1 TGAACTCTGGCCT | Intron1–2 | |
| DR1 TGACCTGTGACCT | Intron1-2 | |
| DR1 TGGCCCCTGACCC | Intron1-2 | |
| DR1 TGACCAGTGAGCT | Intron1-2 | |
| DR1 AGGTGAGAGGGCA | Intron1-2 | |
| DR1 TGACTTCAGACCT | Intron1-2 | |
| Npt1 | DR1 CCACCTCTGACCT | −893 bp |
| Sat1 | DR1 TGTCCCCTGACCT | −908 bp |
| Svct1 | DR1 AGTGCATAGTCCA | −781 bp |
| DR2 TGAACTCGTGACCC | −1824 bp | |
| Mrp6 | DR1 TTCCCTGTGACCT | −1093 bp |
| DR1 TGGACCTTGCCCT | −104 bp | |
| Mate1 | DR1 TGTCCCTTGAACC | −654 bp |
| CAR | DR1 TGTCCTCTGATCT | −399 bp |
Note. The DR1/DR2 sites are predicted by NHRSCAN, a web-based computer program for the prediction of nuclear receptor–binding sites.
Loss of HNF4a in hepatocytes causes marked alterations in gene expression and disturbances in hepatic metabolism of fatty acids, bile acids, and amino acids (Hayhurst et al., 2001; Inoue et al., 2002, 2004). AhR, CAR, PXR, PPARα, Nrf2, FXR, GR, HNF1, C/EBPs, and NF-κB are nuclear receptors/TFs that play important roles in regulating hepatic expression of drug-processing genes (Klaassen and Slitt, 2005; Pascussi et al., 2008). Except for the loss of CAR, hepatic expression/signaling of AhR, PXR, PPARα, Nrf2, FXR, GR, HNF1, C/EBPs, and NF-κB was either unchanged or moderately altered in HNF4a-HNull mice (Fig. 7).
It is of interest that the expression of CAR is dependent on HNF4a in both fetal and adult liver, whereas the expression of PXR is dependent on HNF4a only in fetal liver but not in adult liver (Kyrmizi et al., 2006). A major difference between adult and fetal liver is the absence of bile acid signaling in fetal liver mediated by FXR, which induces PXR gene expression (Jung et al., 2006). In contrast, hepatic expression of CAR is induced in FXR-null mice (Guo et al., 2003). FXR is likely activated in HNF4a-HNull mice because of markedly elevated unconjugated bile acids (Inoue et al., 2004, 2006) as activators of FXR. Activation of FXR may help maintain hepatic expression of PXR in adult HNF4a-HNull mice (Fig. 8). The higher DNA binding of PXR in HNF4a-Hnull mice (Fig. 7C) suggests that PXR is also activated, likely because of elevated bile acids, which are PXR activators (Schuetz et al., 2001).
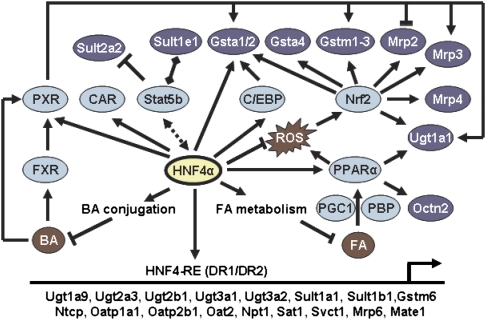
FIG. 8.
Thematic model illustrating the hypothesized direct and indirect roles of HNF4a in regulating hepatic expression of phase II enzymes and transporters in mice. BA, bile acids; FA, fatty acids; HNF4-RE, HNF4a response element; ROS, reactive oxygen species.
In addition to Cyp4a14, hepatic mRNA expression of other PPARα target genes, such as carnitoyl-palmitoyl transferase-II, midchain acyl-CoA dehydrogenase, and 3-hydroxy-3-methylglutaryl CoA synthase, are higher in HNF4a-HNull mice than WT mice (Hayhurst et al., 2001). However, hepatic PPARα mRNA was slightly lower, and the binding of PPAR response elements was also decreased in HNF4a-Hnull mice (Fig. 7C). Currently, the mechanism of hepatic induction of many PPARα target genes in HNF4a-Hnull mice remains unclear. Hepatic accumulation of fatty acids (Hayhurst et al., 2001) and induction of PPARα coactivators, namely PPARγ coactivator 1α (Wiwi et al., 2004) and PBP (Figure 7) in HNF4a-HNull mice may activate PPARα. Interestingly, PPARα and its coactivator PPARγ coactivator 1α can activate PPARα target gene through binding to different gene elements (Song et al., 2010). Therefore, it is likely that induction of PPARα target gene requires the increased interaction of PPARα with its coactivators but might not require an increase of the amount of binding of PPARα to its cis-response elements.
The induction of Nrf2 mRNA and the Nrf2-target genes Ho-1 and Nqo1 indicates that Nrf2 is activated in livers of HNF4a-HNull mice, probably because of increased oxidative stress caused by markedly elevated unconjugated bile acids (Hayhurst et al., 2001; Inoue et al., 2004). Nrf2 is activated in liver during cholestasis (Aleksunes et al., 2006). Alterations in hepatic activities of other TFs may contribute to the complicated patterns of hepatic gene expression in HNF4a-Hnull mice.
OAT2 is the major OAT isoform expressed in human and mouse livers (Buist and Klaassen, 2004) and thus is considered a key transporter in hepatic handling of small organic anions (Rizwan and Burckhardt, 2007). Hepatic and renal expression of Oat2 is lost in HNF1a-null mice (Maher et al., 2006) as well as HNF4a-HNull mice (Fig. 1). Given that hepatic HNF1a mRNA remains largely unchanged in HNF4a-null livers (Fig. 7), it appears that both HNF1a and HNF4a are required for hepatic expression of Oat2 in mice.
The marked downregulation of Oatp1a1 in both male and female HNF4a-null livers indicates that HNF4a is required for hepatic expression of Oatp1a1. The female-predominant hepatic expression of Oatp1a4 in mice is because of suppression by androgens and male-pattern GH secretion (Cheng et al., 2006). Thus, the male-selective induction of Oatp1a4 in HNF4a-null livers may be because of decreased male hormone signaling in these mice, as indicated by the dramatic renal downregulation of Oatp1a1 (Fig. 6), an androgen-activated gene (Cheng et al., 2006), in male HNF4a-null mice. Liver-specific expression of Oatp1b2 requires HNF1a (Maher et al., 2006). The moderate downregulation of Oatp1b2 in HNF4a-null livers is consistent with the moderate decrease of DNA binding of HNF1 in these mice (Fig. 7C).
Little is known about regulation of the Oatp2b1 gene, which is expressed in diverse tissues in mice (Cheng et al., 2005). Although no DR1 or DR2 sites were found in the 2-kb proximal promoter of Oatp2b1, seven DR1 sites were identified in the first intron of mouse Oatp2b1 gene (Table 1), suggesting that HNF4a may bind to these DR1 sites to maintain hepatic expression of Oatp2b1.
Octn2 is critical for cellular uptake of carnitine, which is required for normal mitochondrial fatty acid oxidation. Octn2 is a PPARα target gene (Eder and Ringseis, 2010). Thus, the higher expression of Octn2 in HNF4a-null livers is likely because of activation of PPARα and/or its coactivator (Fig. 8).
The present data demonstrate a critical role of HNF4a in regulating hepatic expression of phosphate and sulfate uptake transporters, Npt1 and Sat1. Npt1 is not only important in the transport of phosphate but also transports various anionic drugs (Yabuuchi et al., 1998). Thus, HNF4a may be important in regulating hepatic uptake of phosphate and certain anionic drugs. The expression of Npt1 is selectively lost in the kidney, but not liver, of HNF1a-null mice (Cheret et al., 2002). Thus, HNF4a and HNF1a are critical for hepatic and renal expression of Npt1, respectively. The sulfate/oxalate exchanger Sat1 is expressed in sinusoidal membranes of hepatocytes (Krick et al., 2009). Loss of Sat1 will likely result in marked decreases in hepatic uptake of sulfate and the sulfation reaction, but accumulation of oxalate in hepatocytes.
The present data indicate that HNF4a is required for hepatic expression of the low-affinity high-capacity vitamin C transporter (Svct1) in mice. An in vitro study suggests that HNF1a is important for hepatic expression of the human SVCT1 gene (Michels and Hagen, 2009). Svct1 is also expressed highly in kidney and intestine (Savini et al., 2008), where HNF4a is enriched. The role of HNF4a in regulating Svct1 expression and its impact on vitamin C homeostasis warrants further investigation.
UGT1A1 is the key enzyme for the glucuronidation of bilirubin, whereas UGT1A9 is important in the detoxification of several carcinogens and clearance of anticancer and pain medications (Olson et al., 2009; Wells et al., 2004). Knockdown of HNF4a in primary human hepatocytes by siRNA results in similar downregulation of UGT1A1 and UGT1A9 (Kamiyama et al., 2007). In the present study, hepatic Ugt1a9 was markedly downregulated, whereas Ugt1a1 was induced in HNF4a-HNull mice (Fig. 1). Hepatic Ugt1a1 mRNA is induced by activators of various nuclear receptors including AhR, CAR, PXR, PPARα, and Nrf2 in mice (Buckley and Klaassen, 2009). Thus, the induction of Ugt1a1 in HNF4a-null livers may be because of the activation of one or multiple nuclear receptors (e.g., PXR and Nrf2). The marked downregulation of Ugt1a9 in HNF4a-null livers is consistent with a previous report (Barbier et al., 2005). These data, in conjunction with a strong correlation of expression of HNF4a with UGT1A9 in human livers (Aueviriyavit et al., 2007), suggest that HNF4a is essential in maintaining hepatic basal expression of UGT1A9 in both humans and mice.
UGT2A3 is highly expressed in human and mouse livers (Buckley and Klaassen, 2007; Court et al., 2008). UGT2A3 selectively glucuronidates bile acids; the most effective substrate is hyodeoxycholic acid, a 6α-hydroxylation detoxification product of lithocholic acid, a toxic secondary bile acid (Court et al., 2008). Little is known about the regulation of Ugt2a3. A critical role for HNF4a in the regulation of Ugt2a3 expression and its impact on hepatic detoxification of lithocholic acid warrants further investigation.
Ugt2b1 has broad substrate specificity, including morphine, phenols, carboxylic acids, and bile acids (e.g., lithocholic acid) (Pritchard et al., 1994; Radominska et al., 1994). In mice, Ugt2b1 is almost exclusively expressed in liver (Buckley and Klaassen, 2007). Mouse Ugt2b1 protein is 80% homologous to human UGT2B4, which is highly expressed in human livers (Izukawa et al., 2009). Hepatic expression of Ugt2b1 requires C/EBPα (Hansen et al., 1998). However, the moderate downregulation of C/EBPα (Wiwi et al., 2004) and decrease of DNA binding of C/EBP proteins (Fig. 7C) alone cannot account for the dramatic loss of Ugt2b1 expression in HNF4a-null livers. Thus, HNF4a is essential for hepatic expression of Ugt2b1 in mice, likely via direct binding to the DR1 site (Table 1) in the proximal promoter of the Ugt2b1 gene.
HNF4a-HNull mice had downregulation of the liver-predominant Ugt2b36. Two DR1 sites were identified in the proximal promoter of Ugt2b36 (Table 1). In contrast, no DR1 sites were found in either the 2-kb proximal promoter or the first intron of the Ugt2b34 and Ugt2b35 gene, whose hepatic expression remains unchanged in HNF4a-HNull mice. Thus, HNF4a is important in regulating hepatic expression of Ugt2b36, most likely via binding to the DR1 sites in the Ugt2b36 gene.
The present data indicate that HNF4a is required for hepatic expression of Ugt3a1 and 3a2 in mice. Ugt3a1 and 3a2 are predominantly expressed in liver and kidney in mice (Buckley and Klaassen, 2007). UGT3A1 is a newly characterized UDP N-acetylglucosaminyltransferase that catalyzes the transfer of N-acetylglucosamine from UDP N-acetylglucosamine to ursodeoxycholic acid and other prototypical UGT1 and UGT2 substrates (Mackenzie et al., 2008).
The present data indicate an important role for HNF4a in the regulation of sulfation reactions. HNF4a is identified as a novel key transactivator of Sult1a1 and Sult1b1 in mouse liver (Fig. 3). Sult1a1 is highly expressed in mouse liver (Alnouti and Klaassen, 2006) and catalyzes the sulfation of phenolic compounds, including steroid hormones, catecholamines, and phenolic drugs, and also participates in the bioactivation of procarcinogens (Hildebrandt et al., 2009). Sult1b1 catalyzes the sulfation of dopa and tyrosine isomers, as well as dopamine and 3,3′,5-triiodo-L-thyronine (Saeki et al., 1998).
A genome-wide study indicates that HNF4a is critical in determining gender-specific gene expression in mouse liver (Holloway et al., 2008). Sult2a1/2 is specifically expressed in female mouse livers (Alnouti and Klaassen, 2006). The high expression of female-specific Sult2a2 mRNA in male HNF4a-null livers is very similar to the high expression of the female-specific Cyp2b9 mRNA in male HNF4a-null livers (Wiwi et al., 2004). Thus, the marked increase of Sult2a2 in HNF4a-null livers may be because of loss of the male-specific inhibition by HNF4a. HNF4a-null livers have marked elevation in unconjugated bile acids because of downregulation of bile acid–conjugating enzymes (Inoue et al., 2004). Sult2a is important in the detoxification of unconjugated bile acids. Induction of Sult2a2 may ameliorate hepatotoxicity induced by the elevated unconjugated bile acids in HNF4a-HNull mice.
The dramatic increase of Sult1e1 mRNA in HNF4a-null livers is very similar to the remarkable increase of Sult1e1 in livers deficient in Stat5b (Holloway et al., 2007), a critical regulator of gender-specific gene expression (Waxman, 2000). Additionally, hepatic Sult1e1 can be markedly induced by dexamethasone through the GR (Alnouti and Klaassen, 2008; Gong et al., 2008). Hepatic basal mRNA expression of Sult1e1 is low in both male and female mice (Alnouti and Klaassen, 2006). Sult1e1 is responsible for the sulfation and inactivation of β-estradiol (E2) at physiological concentrations. Sult1e1 overexpression in hepatoma cells decreased markedly the phosphorylation and activation of Stat5b stimulated by growth hormone (GH) or GH plus E2 (Li et al., 2009). Interestingly, a majority of the sex-specific genes respond similarly to the loss of Stat5b and the loss of HNF4a, indicating that both TFs are essential and they may coregulate sexually dimorphic hepatic gene expression (Holloway et al., 2006); however, the underlying mechanism remains unclear. In fact, HNF4a has been shown to strongly inhibit the phosphorylation of Stat5b and its transcriptional activity (Park et al., 2006). The mechanism of the dramatic induction of Sult1e1 in HNF4a-null livers and its contribution to alterations in Stat5b signaling and gender-divergent gene expression warrant further investigation.
Little is known regarding the regulation of Gsts by HNF4a. It is known that multiple factors are involved in the regulation of a given gene. Interestingly, an HNF4a-like binding site has been identified within 1 kb upstream of the Gsta1 (Gst ya) gene (Paulson et al., 1990). We found two putative DR1 sites within 2 kb of the Gsta1 promoter (DR1 AGATGAAAGTTCA at −1769 bp and DR1 TGCACTGAGACCT at −646 bp). Additionally, we also identified three DR1 sites in intron1-2 of Gsta4 (data not shown) and one DR1 site in the Gstm6 promoter (Table 1). Thus, we speculate that HNF4a might induce the basal expression of Gsta1, Gsta4, and Gstm6. In contrast to the robust induction of Gsta1, Gsta4, and Gstm1-4 by Nrf2 activators, Gstm6 is not consistently induced by Nrf2 activation (Knight et al., 2008). Interestingly, C/EBPβ is essential for the induction of Gsta2 by oltipraz, a dual activator of C/EBPβ and Nrf2 (Ko et al., 2006). Consistent with ∼50% downregulation of C/EBPα and C/EBPβ in HNF4a-Hnull mice (Wiwi et al., 2004), hepatic TF binding to the C/EBP consensus elements decreased ∼50% (Fig. 7C). Additionally, multiple Gsts, including Gsta1/2 and Gstm1-3, can be induced by ligands of PXR (Knight et al., 2008), which appears to be activated in HNF4a-Hnull mice (Fig. 7C). Thus, it is likely that hepatic expression of Gsta1/2 is positively regulated by HNF4a, C/EBP, PXR, and Nrf2. The final effect of HNF4a deficiency on hepatic Gst mRNA expression is likely determined by the combination of vanished transactivation by HNF4a and/or C/EBP but enhanced transactivation by PXR and/or Nrf2. The lack of alteration in Gsta1/Gsta2 mRNA might be because of the neutralizing effects among vanished HNF4a and C/EBP but enhanced PXR and Nrf2. The induction of Gsta4, Gstm1, Gstm2, and Gstm3 in HNF4a-HNull mice is likely because of activation of Nrf2 and PXR. In contrast, a loss of transactivation by HNF4a might contribute to the downregulation of Gstm6 in HNF4a-HNull mice. Further experiments are needed to test the hypothesis that HNF4a might directly regulate Gsta1, Gsta4, Gstm4, and Gstm6 expression in mouse liver.
Mrp family members are important for the efflux of xenobiotics and their conjugated metabolites into bile (by Mrp2) or back into blood (by Mrp3 and Mrp4). The marked decrease of Mrp6 expression in HNF4a-null livers is consistent with an in vitro study that HNF4a binds to the proximal promoter of mouse Mrp6 gene and transactivates Mrp6 expression (Douet et al., 2006). The induction of Mrp3 and Mrp4 in HNF4a-null livers is likely because of activation of PXR and Nrf2, transactivators of mouse Mrp3 and/or Mrp4 (Maher et al., 2007). Activation of Nrf2 induces Mrp2, whereas activation of PXR downregulates Mrp2 in mouse livers (Maher et al., 2007), which might explain the unchanged hepatic Mrp2 expression in HNF4a-Hnull mice (Fig. 5).
Hepatic mRNA expression of the canalicular efflux transporter Bcrp is male predominant in mice because of the inductive effect of testosterone (Tanaka et al., 2005). Thus, the male-selective downregulation of Bcrp in HNF4a-null livers is likely because of a decrease of androgen signaling.
Among the efflux transporters examined, Mate1 is the only transporter whose expression is lost in livers of HNF4a-null mice. Mate1 is important for excretion of organic cations by kidney and liver; renal secretion of metformin is markedly decreased in Mate1-null mice (Tsuda et al., 2009). The role of HNF4a in regulating hepatic and renal expression of Mate1 and its impact on the disposition of cationic drugs remains to be elucidated.
In summary, the present study demonstrates that HNF4a is critical in regulating hepatic mRNA expression of a large number of phase II enzymes and transporters. Although protein levels of these genes were not determined because of the large scope of this study, many of the mRNA level changes are so marked in magnitude that it is likely to have a major impact on protein. The marked alterations in hepatic mRNA expression of these genes is likely because of the loss of HNF4a as a direct transactivator (via binding to the DR1 sites), the loss of gender-specific gene expression (via cross talk with Stat5b), and/or the adaptive activation of certain xenobiotic receptors, such as PXR and Nrf2 (Fig. 8).
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (grants ES-009649, ES-009716, ES-013714, ES-019487, DK081461, and RR021940).
Supplementary Material
Acknowledgments
We thank Cheryl Rockwell, Rachel Chennault, and Xiaohong Lei for technical assistance and the postdoctoral fellows and graduate students of Dr Klaassen's laboratory for critical review of the manuscript.
References
- Aleksunes LM, Slitt AL, Maher JM, Dieter MZ, Knight TR, Goedken M, Cherrington NJ, Chan JY, Klaassen CD, Manautou JE. Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell Stress Chaperones. 2006;11:356–363. doi: 10.1379/CSC-217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol. Sci. 2006;93:242–255. doi: 10.1093/toxsci/kfl050. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J. Pharmacol. Exp. Ther. 2008;324:612–621. doi: 10.1124/jpet.107.129650. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Petrick JS, Klaassen CD. Tissue distribution and ontogeny of organic cation transporters in mice. Drug Metab. Dispos. 2006;34:477–482. doi: 10.1124/dmd.105.006932. [DOI] [PubMed] [Google Scholar]
- Aueviriyavit S, Furihata T, Morimoto K, Kobayashi K, Chiba K. Hepatocyte nuclear factor 1 alpha and 4 alpha are factors involved in interindividual variability in the expression of UGT1A6 and UGT1A9 but not UGT1A1, UGT1A3 and UGT1A4 mRNA in human livers. Drug Metab. Pharmacokinet. 2007;22:391–398. doi: 10.2133/dmpk.22.391. [DOI] [PubMed] [Google Scholar]
- Barbier O, Girard H, Inoue Y, Duez H, Villeneuve L, Kamiya A, Fruchart JC, Guillemette C, Gonzalez FJ, Staels B. Hepatic expression of the UGT1A9 gene is governed by hepatocyte nuclear factor 4alpha. Mol. Pharmacol. 2005;67:241–249. doi: 10.1124/mol.104.003863. [DOI] [PubMed] [Google Scholar]
- Berasain C, Herrero JI, Garcia-Trevijano ER, Avila MA, Esteban JI, Mato JM, Prieto J. Expression of Wilms' tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology. 2003;38:148–157. doi: 10.1053/jhep.2003.50269. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab. Dispos. 2007;35:121–127. doi: 10.1124/dmd.106.012070. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab. Dispos. 2009;37:847–856. doi: 10.1124/dmd.108.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist SC, Klaassen CD. Rat and mouse differences in gender-predominant expression of organic anion transporter (Oat1-3; Slc22a6-8) mRNA levels. Drug Metab. Dispos. 2004;32:620–625. doi: 10.1124/dmd.32.6.620. [DOI] [PubMed] [Google Scholar]
- Cheng X, Buckley D, Klaassen CD. Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem. Pharmacol. 2007;74:1665–1676. doi: 10.1016/j.bcp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. Regulation of mRNA expression of xenobiotic transporters by the pregnane x receptor in mouse liver, kidney, and intestine. Drug Metab. Dispos. 2006;34:1863–1867. doi: 10.1124/dmd.106.010520. [DOI] [PubMed] [Google Scholar]
- Cheng X, Maher J, Chen C, Klaassen CD. Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps) Drug Metab. Dispos. 2005;33:1062–1073. doi: 10.1124/dmd.105.003640. [DOI] [PubMed] [Google Scholar]
- Cheng X, Maher J, Lu H, Klaassen CD. Endocrine regulation of gender-divergent mouse organic anion-transporting polypeptide (Oatp) expression. Mol. Pharmacol. 2006;70:1291–1297. doi: 10.1124/mol.106.025122. [DOI] [PubMed] [Google Scholar]
- Cheret C, Doyen A, Yaniv M, Pontoglio M. Hepatocyte nuclear factor 1 alpha controls renal expression of the Npt1-Npt4 anionic transporter locus. J. Mol. Biol. 2002;322:929–941. doi: 10.1016/s0022-2836(02)00816-1. [DOI] [PubMed] [Google Scholar]
- Court MH, Hazarika S, Krishnaswamy S, Finel M, Williams JA. Novel polymorphic human UDP-glucuronosyltransferase 2A3: cloning, functional characterization of enzyme variants, comparative tissue expression, and gene induction. Mol. Pharmacol. 2008;74:744–754. doi: 10.1124/mol.108.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab. Dispos. 2009;37:203–210. doi: 10.1124/dmd.108.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet V, VanWart CM, Heller MB, Reinhard S, Le Saux O. HNF4alpha and NF-E2 are key transcriptional regulators of the murine Abcc6 gene expression. Biochim. Biophys. Acta. 2006;1759:426–436. doi: 10.1016/j.bbaexp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder K, Ringseis R. The role of peroxisome proliferator-activated receptor alpha in transcriptional regulation of novel organic cation transporters. Eur. J. Pharmacol. 2010;628:1–5. doi: 10.1016/j.ejphar.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Gong H, Jarzynka MJ, Cole TJ, Lee JH, Wada T, Zhang B, Gao J, Song WC, DeFranco DB, Cheng SY, et al. Glucocorticoids antagonize estrogens by glucocorticoid receptor-mediated activation of estrogen sulfotransferase. Cancer Res. 2008;68:7386–7393. doi: 10.1158/0008-5472.CAN-08-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab. Pharmacokinet. 2008;23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Hansen AJ, Lee YH, Sterneck E, Gonzalez FJ, Mackenzie PI. C/EBPalpha is a regulator of the UDP glucuronosyltransferase UGT2B1 gene. Mol. Pharmacol. 1998;53:1027–1033. [PubMed] [Google Scholar]
- Hartley DP, Klaassen CD. Detection of chemical-induced differential expression of rat hepatic cytochrome P450 mRNA transcripts using branched DNA signal amplification technology. Drug Metab. Dispos. 2000;28:608–616. [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M, Adjei A, Weinshilboum R, Johnson JA, Berlin DS, Klein TE, Altman RB. Very important pharmacogene summary: sulfotransferase 1A1. Pharmacogenet. Genomics. 2009;19:404–406. doi: 10.1097/FPC.0b013e32832e042e. [DOI] [PubMed] [Google Scholar]
- Holloway MG, Cui Y, Laz EV, Hosui A, Hennighausen L, Waxman DJ. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148:1977–1986. doi: 10.1210/en.2006-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway MG, Laz EV, Waxman DJ. Codependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4alpha. Mol. Endocrinol. 2006;20:647–660. doi: 10.1210/me.2005-0328. [DOI] [PubMed] [Google Scholar]
- Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol. Endocrinol. 2008;22:1274–1286. doi: 10.1210/me.2007-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ. Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4alpha (HNF4alpha). HNF4alpha regulates ornithine transcarbamylase in vivo. J. Biol. Chem. 2002;277:25257–25265. doi: 10.1074/jbc.M203126200. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte nuclear factor 4alpha is a central regulator of bile acid conjugation. J. Biol. Chem. 2004;279:2480–2489. doi: 10.1074/jbc.M311015200. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, Inoue J, Xiang CC, Brownstein MJ, Eggertsen G, Bjorkhem I, et al. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J. Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, Aoki Y, Ikushiro S, Sakaki T, Yokoi T. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab. Dispos. 2009;37:1759–1768. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- Jover R, Moya M, Gomez-Lechon MJ. Transcriptional regulation of cytochrome p450 genes by the nuclear receptor hepatocyte nuclear factor 4-alpha. Curr. Drug Metab. 2009;10:508–519. doi: 10.2174/138920009788898000. [DOI] [PubMed] [Google Scholar]
- Jung D, Mangelsdorf DJ, Meyer UA. Pregnane X receptor is a target of farnesoid X receptor. J. Biol. Chem. 2006;281:19081–19091. doi: 10.1074/jbc.M600116200. [DOI] [PubMed] [Google Scholar]
- Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab. Pharmacokinet. 2007;22:287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
- Kang X, Zhong W, Liu J, Song Z, McClain CJ, Kang YJ, Zhou Z. Zinc supplementation reverses alcohol-induced steatosis in mice through reactivating hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Hepatology. 2009;50:1241–1250. doi: 10.1002/hep.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Niehof M, Matys V, Zemlin R, Borlak J. Genome wide prediction of HNF4alpha functional binding sites by the use of local and global sequence context. Genome Biol. 2008;9:R36. doi: 10.1186/gb-2008-9-2-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr. Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. Induction of hepatic glutathione S-transferases in male mice by prototypes of various classes of microsomal enzyme inducers. Toxicol. Sci. 2008;106:329–338. doi: 10.1093/toxsci/kfn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MS, Lee SJ, Kim JW, Lim JW, Kim SG. Differential effects of the oxidized metabolites of oltipraz on the activation of CCAAT/enhancer binding protein-beta and NF-E2-related factor-2 for GSTA2 gene induction. Drug Metab. Dispos. 2006;34:1353–1360. doi: 10.1124/dmd.106.009514. [DOI] [PubMed] [Google Scholar]
- Krick W, Schnedler N, Burckhardt G, Burckhardt BC. Ability of sat-1 to transport sulfate, bicarbonate, or oxalate under physiological conditions. Am. J. Physiol. Renal Physiol. 2009;297:F145–F154. doi: 10.1152/ajprenal.90401.2008. [DOI] [PubMed] [Google Scholar]
- Kuo SM, MacLean ME, McCormick K, Wilson JX. Gender and sodium-ascorbate transporter isoforms determine ascorbate concentrations in mice. J. Nutr. 2004;134:2216–2221. doi: 10.1093/jn/134.9.2216. [DOI] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology. 2004;39:1038–1047. doi: 10.1002/hep.20155. [DOI] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Li L, He D, Wilborn TW, Falany JL, Falany CN. Increased SULT1E1 activity in HepG2 hepatocytes decreases growth hormone stimulation of STAT5b phosphorylation. Steroids. 2009;74:20–29. doi: 10.1016/j.steroids.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie PI, Rogers A, Treloar J, Jorgensen BR, Miners JO, Meech R. Identification of UDP glycosyltransferase 3A1 as a UDP N-acetylglucosaminyltransferase. J. Biol. Chem. 2008;283:36205–36210. doi: 10.1074/jbc.M807961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Callaghan TN, Cheng X, Cheung C, Gonzalez FJ, Klaassen CD. Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1alpha. Biochem. Pharmacol. 2006;72:512–522. doi: 10.1016/j.bcp.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Cherrington NJ, Cheng X, Klaassen CD. Tissue distribution and hepatic and renal ontogeny of the multidrug resistance-associated protein (Mrp) family in mice. Drug Metab. Dispos. 2005;33:947–955. doi: 10.1124/dmd.105.003780. [DOI] [PubMed] [Google Scholar]
- Markovich D, Aronson PS. Specificity and regulation of renal sulfate transporters. Annu. Rev. Physiol. 2007;69:361–375. doi: 10.1146/annurev.physiol.69.040705.141319. [DOI] [PubMed] [Google Scholar]
- Mazure NM, Nguyen TL, Danan JL. Severe hypoxia specifically downregulates hepatocyte nuclear factor-4 gene expression in HepG2 human hepatoma cells. Tumour Biol. 2001;22:310–317. doi: 10.1159/000050632. [DOI] [PubMed] [Google Scholar]
- Michels AJ, Hagen TM. Hepatocyte nuclear factor 1 is essential for transcription of sodium-dependent vitamin C transporter protein 1. Am. J. Physiol. Cell Physiol. 2009;297:C1220–C1227. doi: 10.1152/ajpcell.00348.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama C, Hi R, Osada S, Osumi T. Functional interactions between nuclear receptors recognizing a common sequence element, the direct repeat motif spaced by one nucleotide (DR-1) J. Biochem. 1998;123:1174–1179. doi: 10.1093/oxfordjournals.jbchem.a022058. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KC, Dellinger RW, Zhong Q, Sun D, Amin S, Spratt TE, Lazarus P. Functional characterization of low-prevalence missense polymorphisms in the UDP-glucuronosyltransferase 1A9 gene. Drug Metab. Dispos. 2009;37:1999–2007. doi: 10.1124/dmd.108.024596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Wiwi CA, Waxman DJ. Signalling cross-talk between hepatocyte nuclear factor 4alpha and growth-hormone-activated STAT5b. Biochem. J. 2006;397:159–168. doi: 10.1042/BJ20060332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Duret C, Daujat-Chavanieu M, Vilarem MJ, Maurel P. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu. Rev. Pharmacol. Toxicol. 2008;48:1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- Paulson KE, Darnell JE, Jr, Rushmore T, Pickett CB. Analysis of the upstream elements of the xenobiotic compound-inducible and positionally regulated glutathione S-transferase Ya gene. Mol. Cell. Biol. 1990;10:1841–1852. doi: 10.1128/mcb.10.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrick JS, Klaassen CD. Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab. Dispos. 2007;35:1806–1815. doi: 10.1124/dmd.107.015974. [DOI] [PubMed] [Google Scholar]
- Pritchard M, Fournel-Gigleux S, Siest G, Mackenzie P, Magdalou J. A recombinant phenobarbital-inducible rat liver UDP-glucuronosyltransferase (UDP-glucuronosyltransferase 2B1) stably expressed in V79 cells catalyzes the glucuronidation of morphine, phenols, and carboxylic acids. Mol. Pharmacol. 1994;45:42–50. [PubMed] [Google Scholar]
- Radominska A, Little JM, Lester R, Mackenzie PI. Bile acid glucuronidation by rat liver microsomes and cDNA-expressed UDP-glucuronosyltransferases. Biochim. Biophys. Acta. 1994;1205:75–82. doi: 10.1016/0167-4838(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Rizwan AN, Burckhardt G. Organic anion transporters of the SLC22 family: biopharmaceutical, physiological, and pathological roles. Pharm. Res. 2007;24:450–470. doi: 10.1007/s11095-006-9181-4. [DOI] [PubMed] [Google Scholar]
- Ryffel GU. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Sakakibara Y, Araki Y, Yanagisawa K, Suiko M, Nakajima H, Liu MC. Molecular cloning, expression, and characterization of a novel mouse liver SULT1B1 sulfotransferase. J. Biochem. 1998;124:55–64. doi: 10.1093/oxfordjournals.jbchem.a022097. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol. Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- Savini I, Rossi A, Pierro C, Avigliano L, Catani MV. SVCT1 and SVCT2: key proteins for vitamin C uptake. Amino Acids. 2008;34:347–355. doi: 10.1007/s00726-007-0555-7. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, et al. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J. Biol. Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
- Song S, Attia RR, Connaughton S, Niesen MI, Ness GC, Elam MB, Hori RT, Cook GA, Park EA. Peroxisome proliferator activated receptor alpha (PPARalpha) and PPAR gamma coactivator (PGC-1alpha) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol. Cell. Endocrinol. 2010;325:54–63. doi: 10.1016/j.mce.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou S, Gispert S, Cheng J, Wang Y, Chen A, Hoogstraten-Miller S, Miller GF, Kwon O, Levine M, Guttentag SH, et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat. Med. 2002;8:514–517. doi: 10.1038/0502-514. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Slitt AL, Leazer TM, Maher JM, Klaassen CD. Tissue distribution and hormonal regulation of the breast cancer resistance protein (Bcrp/Abcg2) in rats and mice. Biochem. Biophys. Res. Commun. 2005;326:181–187. doi: 10.1016/j.bbrc.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui K. Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol. Pharmacol. 2009;75:1280–1286. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- Waxman DJ. Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression. Novartis Found. Symp. 2000;227:61–74. doi: 10.1002/0470870796.ch5. discussion 75–81. [DOI] [PubMed] [Google Scholar]
- Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, Hansen AJ, Kessler FK, Kim PM, Chowdhury NR, et al. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab. Dispos. 2004;32:281–290. doi: 10.1124/dmd.32.3.281. [DOI] [PubMed] [Google Scholar]
- Wiwi CA, Gupte M, Waxman DJ. Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4alpha-deficient mice. Mol. Endocrinol. 2004;18:1975–1987. doi: 10.1210/me.2004-0129. [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab. Dispos. 2007;35:1700–1710. doi: 10.1124/dmd.107.016436. [DOI] [PubMed] [Google Scholar]
- Yabuuchi H, Tamai I, Morita K, Kouda T, Miyamoto K, Takeda E, Tsuji A. Hepatic sinusoidal membrane transport of anionic drugs mediated by anion transporter Npt1. J. Pharmacol. Exp. Ther. 1998;286:1391–1396. [PubMed] [Google Scholar]
- Yaoi T, Jiang X, Li X. Development of a fluorescent microsphere-based multiplexed high-throughput assay system for profiling of transcription factor activation. Assay Drug Dev. Technol. 2006;4:285–292. doi: 10.1089/adt.2006.4.285. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Kang X, Jiang Y, Song Z, Feng W, McClain CJ, Kang YJ. Preservation of hepatocyte nuclear factor-4alpha is associated with zinc protection against TNF-alpha hepatotoxicity in mice. Exp. Biol. Med. 2007;232:622–628. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.