Abstract
Exposure during the organogenesis stage of the mouse embryo to the model teratogen, hydroxyurea (HU), induces curly tail and limb malformations. Oxidative stress contributes to the developmental toxicity of HU. Reactive oxygen species (ROS) interact with polyunsaturated bilipid membranes to form α,β-unsaturated reactive aldehydes; 4-hydroxy-2-nonenal (4-HNE), one of the most cytotoxic of these aldehydes, covalently adducts with proteins, lipids, and nucleic acids. The goal of the current study is to determine if HU exposure of CD1 mice on gestation day 9 generates region-specific 4-HNE-protein adducts in the embryo and to identify the proteins targeted. The formation of 4-HNE-protein adducts was elevated in the caudal region of control embryos; HU exposure further increased 4-HNE-protein adduct formation in this area. Interestingly, three of the 4-HNE–modified proteins, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glutamate oxaloacetate transaminase 2, and aldolase 1, A isoform, are involved in energy metabolism. The formation of 4-HNE-GAPDH protein adducts reduced GAPDH enzymatic activity by 20% and attenuated lactate production by 40%. Furthermore, HU exposure induced the nuclear translocation of GAPDH in the caudal region of exposed embryos; this nuclear translocation may be associated with the reactivation of oxidized proteins involved in DNA repair, such as apurinic/apyrimidinic endonuclease-1, and the stimulation of E1A-associated P300 protein/creb-binding protein (p300/CBP) activity, initiating cell death in a p53-dependent pathway. We propose that GAPDH is a redox-sensitive target in the embryo and may play a role in a stress response during development.
Keywords: 4-hydroxy-2-nonenal, protein adducts, glyceraldehyde-3-phosphate dehydrogenase, oxidative stress, nuclear translocation, glycolysis
Reactive oxygen species (ROS) play a significant role in embryonic development. ROS modulate signaling pathways that promote differentiation, proliferation, and apoptosis (Dennery, 2007). Excess ROS lead to oxidative stress and the perturbation of redox-sensitive signaling pathways, some of which are associated with dysmorphogenesis (Ozolins and Hales, 1997). During midorganogenesis, a period when the embryo is undergoing rapid cellular growth and differentiation leading to major structural changes, ROS may play an important role in mediating teratogenic insult. The mechanisms by which oxidative stress leads to embryotoxicity and teratogenesis are still not fully understood.
ROS contribute to embryonic maldevelopment by modifying redox-sensitive signaling pathways critical to development. During organogenesis, oxidative stress posttranslationally modifies redox-regulated transcription factors with subsequent changes in gene expression in the embryo (Dennery, 2007). A caudal isoform of Wnt, Wnt8c, important for body patterning and organogenesis (Geetha-Loganathan, 2008), is perturbed by H2O2, inducing rapid stabilization of β-catenin and increasing the expression of Wnt target genes (Korswagen, 2006). Activator protein-1 (AP-1) is a redox-sensitive early response transcription factor composed of Jun and Fos families of nuclear proteins whose gene products are associated with antioxidant defense, cell cycle control, and apoptosis. Hydroxyurea (HU), a free-radical producing teratogen, induces oxidative stress during organogenesis, enhancing the formation of c-Fos heterodimers and leading to increased AP-1 DNA-binding activity (Yan and Hales, 2005). Transgenic overexpression of superoxide dismutase (hSOD1) in dams treated with HU lowers the incidence of malformations (Larouche and Hales, 2009). It is clear that oxidative stress plays a critical role in HU-induced developmental defects.
ROS may also cause embryotoxicity by inducing lipid peroxidation. Free-radical chain oxidation reactions with polyunsaturated fatty acids within cellular membranes produce electrophilic aldehydes (Sayre et al., 2006). 4-Hydroxy-2-nonenal (4-HNE) is one of the most widely recognized and most studied cytotoxic products of lipid peroxidation (Uchida, 2003). 4-HNE is diffusible; it may act locally or be distributed to other tissues through the circulation (Sayre et al., 2006). Michael addition reactions with the side chain of cysteine are highly susceptible to covalent modifications by 4-HNE (Grimsrud et al., 2008). Covalent adducts of 4-HNE are key regulators of the stress response. A number of the protein targets of 4-HNE affect energy production. Glycolytic enzymes, namely, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphofructokinase, aldolase, and lactate dehydrogenase, are redox sensitive and form covalent modifications with 4-HNE (Novotny et al., 1994). Previous studies from our laboratory have revealed the formation of region-specific 4-HNE adducts in teratogen-exposed organogenesis stage mouse embryos.
Glycolytic enzymes are of particular interest as sensitive targets of ROS within the developing embryo because they are critically required for ATP production (Dumollard et al., 2009; Grant, 2008; Neubert, 1970). GAPDH is inactivated during periods of oxidative stress and is modified by 4-HNE in several experimental models (Botzen and Grune, 2007; Schutt et al., 2003; Uchida and Stadtman, 1993). GAPDH, once thought to be involved only in cellular maintenance, is now recognized as an important multifunctional entity. There is compelling evidence that GAPDH is a developmentally critical enzyme that protects the embryo from endogenous and xenobiotic-initiated oxidative stress and DNA damage that results in a broad range of embryopathies. Posttranslational modifications of GAPDH during oxidative stress are thought to be responsible for its multiple cellular functions, including p53-dependent cell death (Hwang et al., 2009; Sen et al., 2008). HU, a model teratogen, is used as an anticancer agent and for the treatment of sickle cell disease. HU is a potent inhibitor of class I ribonucleotide reductase, quenching the tyrosyl free radical within its catalytic center, resulting in inhibition of DNA replication and the induction of oxidative stress. HU exposure on gestation day 9 (GD 9) causes a dose-dependent increase in caudal defects, mainly hindlimb and lumbosacral vertebral column defects. Pretreatment with antioxidants ameliorates HU-induced malformations and impedes the onset of apoptosis (DeSesso, 1981; DeSesso et al., 1994). We have shown previously that HU causes a dose-dependent increase in 4-HNE-protein adduct immunoreactivity in the caudal tissues in organogenesis stage embryos; HU-induced malformations and 4-HNE adducts were increased by treatment with L-buthionine-S,R-sulfoximine, an irreversible inhibitor of γ-glutamylcysteine synthetase that results in the depletion of glutathione and an increase in oxidative stress (Yan and Hales, 2006). The role of 4-HNE-protein adducts in oxidative stress–induced teratogenesis and the region-specific effects and pathways abrogated by 4-HNE that have a potential impact on caudal malformations have yet to be defined. The goal of this study is to determine if HU exposure generates 4-HNE-protein adducts in malformation-sensitive regions of the embryo, identify the proteins targeted, assess the effects on protein function, and localize 4-HNE-adducted targets.
MATERIALS AND METHODS
Timed-pregnant CD1 mice were purchased from Charles River Canada Ltd (St Constant, Quebec, Canada) and housed in the McIntyre Animal Resource Centre (McGill University, Montreal, Quebec, Canada). Animal protocols were conducted in accordance with the guidelines outlined in the Guide to the Care and Use of Experimental Animals. CD1 mice mated between 8:00 and 10:00 A.M. (GD 0) were treated with saline (control) or HU (Aldrich Chemical Co., Madison, WI) at 400 or 600 mg/kg by ip injection at 9:00 a.M. on GD 9. Female mice were euthanized by CO2 and cervical dislocation on GD 9 at 3 h after treatment; each treatment group consisted of 7–12 litters. On GD 9, the uteri were removed and embryos were explanted to Hanks’ balanced salt solution (Invitrogen Canada, Inc., Ontario, Canada). Embryos were cut into head, body, and tail sections (Fig. 1). Protein extracts were obtained immediately for assessment of 4-HNE-protein adducts by Western blotting or the analysis of GAPDH enzymatic activity. For two-dimensional (2D) gel electrophoresis, the tail parts from four litters of embryos exposed to vehicle or HU600 were pooled and subjected to protein extraction immediately. Embryos were left whole for lactate assays and immunofluorescence.
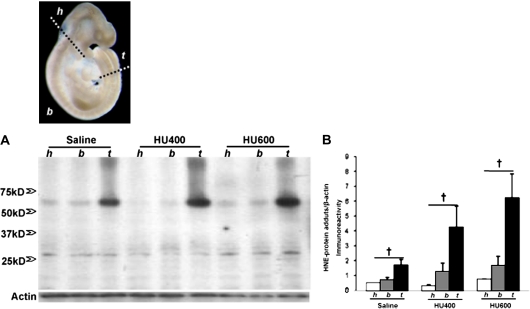
FIG. 1.
Illustration of the separation of the mouse embryo. Head part (h), from the cranial end (top) of the embryo to the caudal end of the first branchial arch; body part (b), the region between the head and the tail part; tail part (t), from the cranial border of the third somites (counted from the caudal end) to the caudal end of the embryo. (A) Western blot analysis of 4-HNE-protein adducts in the three parts of embryos exposed to vehicle (saline) or HU (HU400, 400 mg/kg or HU600, 600 mg/kg). All 4-HNE-protein adducts were quantified by scan densitometric analysis, as indicated in (B). Each bar (mean ± SEM) represents three litters. “Dagger” denotes a significant difference between different parts of the embryo within the same treatment group (†p < 0.05).
1D and 2D gel electrophoresis and Western blot analysis of 4-HNE-protein adducts.
For 4-HNE-protein adduct determination using 1D gel electrophoresis, 7.5 μg of protein from each sample was separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) (15 wells) and then transferred onto equilibrated polyvinylidene difluoride membranes (PVDF) (Amersham Biosciences, Buckinghamshire, U.K.) by electroblotting. Protein concentrations were determined using the Bio-Rad Bradford protein assay (Bio-Rad Laboratories, Ontario, Canada). To analyze GAPDH using Western blotting of proteins from fractionated (head, body, and tail) embryos, 20 μg of protein from each sample was separated by 8% SDS-PAGE and then transferred onto PVDF membranes (Invitrogen Corporation). Membranes were blocked using 10% nonfat milk and then probed with primary antibodies against 4-HNE-protein adducts (1:500; OXIS International, Inc., Foster City, CA), GAPDH (1:1000; Abcam, Inc., Cambridge, MA), and β-actin (1:5000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) overnight at 4°C. Membranes were incubated with horseradish peroxidase–conjugated secondary antibodies (1:10,000; GE Healthcare, Buckinghamshire, U.K.) for 2 h at room temperature, and proteins were detected by enhanced chemiluminescence (GE Healthcare). Protein bands were quantified by densitometric analysis using a ChemiImager 400 Imaging system (Alpha Innotech, San Leandro, CA); the peak area represents the intensity of the signal.
For 2D gel electrophoresis, protein determination, separation, and mass spectrometry were conducted by the Genome Quebec Innovation Centre (Montreal, Quebec, Canada). Protein concentration was determined with use of 2D Quant Kit (Amersham, Baie D'Urfe, Quebec, Canada). Fifty micrograms of protein in 155 μl of Destreak rehydration buffer supplemented with 1% IPG Buffer (3–10 NL, Amersham) were placed in each of four chambers of ZOOM IPGRunner Cassette (Invitrogen). ZOOM Dry Strips (7 cm, pH range 3–10 NL, Invitrogen) were placed in the chambers that were then sealed and left for rehydration for 16 h. The cassette was inserted in ZOOM IPGRunner (Invitrogen Canada, Inc.) and voltage gradient (200–2000 V) was applied as recommended by the manufacturer. A total of 2000 Vh was applied. After isoelectric focusing was completed, strips were equilibrated with SDS using NuPAGE LDS Sample Buffer (1×, Invitrogen Canada, Inc.), reduced with 2% dithiothreitol (DTT, Amersham), and alkylated with iodacetamide (IAA, Sigma). Both steps (DTT and IAA) were done at room temperature for 15 min. Electrophoresis in the second dimension (SDS-PAGE) was done on gradient 4–12% BisTris precast minigels (Invitrogen) immobilized with 1% agarose in XCell SureLock (Invitrogen). The upper and lower chambers were filled with MOPS Running Buffer (Invitrogen); molecular weight standards (Broad Range Protein Molecular Weight Markers, Amersham, 0.9 μg/gel) were placed in the marker wells, and 200 V were applied for 50 min. After completion of the run, one of every pair of gels was fixed overnight in 50% methanol/10% acetic acid fixation solution and stained with silver nitrate by modified Shevchenko's protocol (Yan et al., 2000). Gel images were acquired on an ImageScanner (Amersham) in TIF format.
Immediately after SDS-PAGE, the second gel was covered with a nitrocellulose membrane, placed between two sheets of Whatman paper and two sponges, and inserted into an XCell II Blot Module (Invitrogen). The module was then placed in XCell SureLock. Transfer Buffer (Invitrogen) supplemented with 10% methanol was added in the module, and protein transfer was carried out at 30 V for 1 h. The quality of the protein transfer was controlled by staining with Ponceau S. The membranes were blocked with 5% skim milk for 1 h at room temperature after washing off the Ponceau S stain and then probed with primary antibodies against 4-HNE–modified proteins (1:500, OXIS International, Inc.) overnight at 4°C. After incubation with horseradish peroxidase–conjugated secondary antibodies (1:10,000) for 2 h at room temperature, proteins were detected with the same method as 1D gels.
Mass spectrometry.
Protein spots on the silver-stained 2D gels that correspond to those detected by immunoblotting of 4-HNE–modified proteins were excised from the gel and subjected to trypsin digestion. Sample injection and high-performance liquid chromatography (HPLC) separation were done using an Agilent 1100 series system (Agilent Technologies, Ontario, Canada). Twenty microliters of digest solution were loaded onto a Zorbax 300SB-C18 5 × 0.3 mm trapping column and washed for 5 min at 15 μl/min with 3% acetonitrile (ACN):0.1% formic acid (FA). Nano-HPLC peptide separation was done using a New Objective (Woburn, MA) Biobasic C18 10 × 0.075 mm picofrit analytical column. The gradient was 10% ACN:0.1% FA to 95% ACN:0.1% FA in 15 min at 200 nl/min.
Mass spectrometry was done with a QTRAP 4000 from Sciex-Applied Biosystems (Concord, Ontario, Canada). Information-dependent mass spectrometry/mass spectrometry (MS/MS) analysis was done on the three most intense ions selected from each full scan MS with dynamic exclusion for 90 s. The survey scan used was an enhanced MS scan from 3500 to 1600 m/z at 4000 amu/s using dynamic fill time. MS/MS data were acquired for three scans from 70 to 1700 m/z using a fixed 25 ms trap fill time and with Q0 trapping activated. Peaklists were generated with Mascot Distiller 1.1 from Matrixscience (Boston, MA). Searches of sequences from NCBInr database using a rodent taxonomy filter (157,986 sequences) were done with Mascot 1.9 using trypsin as digestion enzyme, carboxyamidomethylation of cysteines as fixed modification, and methionine oxidation as variable modification and 1.5 Da precursor and 0.8 fragment search tolerances.
2D Western blot analysis of GAPDH.
Membranes obtained from 2D Western blot of 4-HNE–modified proteins were stripped once with stripping buffer (2% SDS; 62.5mM Tris, pH 6.7; 100mM β-mercaptoethanol). After washing three times with tris-buffered saline tween-20 (0.1% Tween-20) for 10 min each, membranes were blocked with 5% skim milk for 1 h at room temperature, probed with a primary antibody against GAPDH (1:2000, rabbit polyclonal immunoglobulin G [IgG], Abcam, Inc.) at 4°C overnight, and incubated with horseradish peroxidase–conjugated secondary antibody (1:10,000, GE Healthcare) for 2 h at room temperature. Proteins were detected and quantified with the same protocol as 1D gels.
GAPDH enzyme assay.
GD 9 embryos were separated into head, body, and tail and pooled from three litters for each treatment group, homogenized in lysis buffer containing protease cocktail inhibitor, and used for the two-step enzymatic reaction in the production of reduced nicotinamide adenine dinucleotide (NADH). The enzyme assay mixture contained 100mM triethanolamine buffer, pH 7.6, 100mM 3-phosphoglyceric acid, 200mM L-cysteine HCL, 100mM magnesium sulfate, 7.0mM β-nicotinamide adenine dinucleotide, 34mM adenosine triphosphate, and 200 units/ml 3-phosphoglyceric phosphokinase (3-PGK); all reagents were dissolved in deionized water. 3-PGK was added last to start the reaction; all reagents were mixed by inversion and equilibrated to 25°C. Spectrophotometric absorbance was measured for NADH production at 340 nm for approximately 5 min, and the ΔA340/min was measured using the maximum linear rate for both test and blank.
Lactate assay.
GD 9 whole embryos were pooled from three litters from each treatment group, homogenized in lysis buffer, flash frozen, and stored at −80°C. Reaction ingredients, including 700 units/l of lactate oxidase, 508 units/l of peroxidase, 2.0 mmol/l of dichlorobenzenesulfonic acid (DCBSA), and 1.16 mmol/l of 4-aminoantipyrine (4-AAP), were added to the homogenized sample (Policy and Procedure Central Laboratory, Biochemistry, McGill University Health Centre). In the reaction, lactate oxidase converts lactate into pyruvate with the simultaneous generation of hydrogen peroxide (H2O2). The H2O2 reacts with DCBSA and 4-AAP to form a chromophore, catalyzed by peroxidase. The lactic acid concentration is determined by measuring the change in absorbance at 520 nm.
Immunofluorescence.
Three hours after the treatment of dams with HU or vehicle on GD 9, embryos were explanted and immersed in 4% paraformaldehyde for 4 h at 4°C. Embryos were then dehydrated in ethanol, embedded in paraffin, and serially sectioned (5-μm sections) along the sagittal plane. Tissues were deparaffinized and rehydrated with PBS for 5 min and then incubated for 30 min in blocking serum (0.1% bovine serum albumin, 0.1% Triton X-100, and PBS). Blocking serum was gently tipped off the slides, and then the sections were incubated with GAPDH primary antibody (1:100; Abcam, Inc., diluted in blocking serum) overnight at 4°C in a humidified chamber. Sections were rinsed three times for 5 min in PBS and then incubated with secondary anti-rabbit IgG (1:200; Vector Laboratories, Inc.) diluted in blocking serum for 1 h at room temperature. After washing three times for 5 min in PBS, tissues were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min and then washed again with PBS for 5 min. Slides were mounted with Vectashield for fluorescence (Vector Laboratories, Inc.). As a negative control, only the secondary antibody was added.
Confocal microscopy and quantitative analysis.
A Zeiss LSM 510 Axiovert 100 M confocal microscope with a Plan-Apochromat ×63/1.4 oil digital image correlation objective was used to visualize the fluorescent GAPDH-stained sagittal sections of embryos treated with vehicle and HU. Optimal settings for laser scanning fluorescence imaging were determined experimentally for both GAPDH antibody and DAPI. All embryos were scanned at a 1.28-μs pixel time speed with an optical slice of approximately 0.6 μm, zoom factor equal to one, and a pinhole setting of 82 μm for GAPDH and 1000 μm for DAPI. Z-stack images of six independent vehicle-treated and seven independent HU600-treated embryos were acquired, and quantitative analysis was done with IMARIS Software (Bitplane AG, Zurich, Switzerland). 3D iso-surfaces of vehicle- and HU600-treated embryos were generated. Only lumbosacral somites were analyzed in this study. 3D surface–rendered GAPDH was isolated from the 3D surface–rendered DAPI. The software enabled GAPDH to be subtracted from the cytoplasm, isolating the colocalized DAPI and GAPDH surfaces to be analyzed by intensity mean.
Statistical analyses.
Statistical analyses were done by a one-way or a two-way ANOVA on ranks, as appropriate, using the SigmaStat computer program, followed by a post hoc Holm-Sidak or Dunn's analysis of the 1D and 2D gel electrophoresis. A one-way ANOVA, followed by the Bonferroni correction for multiple testing, was done for the GAPDH enzymatic analysis and lactate measurements. A Student’s t-test was performed using GraphPrism to test for intensity mean differences. The α priori level of significance was p < 0.05.
RESULTS
4-HNE Is Differentially Localized in the Embryo
Proteins extracted from head, body, and tail of control and HU-exposed embryos were analyzed by Western blot analysis to determine the relative distribution of 4-HNE-protein adducts. A major 4-HNE immunoreactive band was observed in the 65 kDa molecular weight range (Fig. 1A). Interestingly, a higher content of the 65 kDa 4-HNE-protein adducts was found in the tail than in the head or body region of both control embryos and those treated with HU (400 or 600 mg/kg) (Fig. 1B). Multiple low-intensity bands of 4-HNE-protein adducts were detected below 50 kDa (Fig. 2A), but none were greater in content in any of the three body regions of the embryo in any treatment group (Fig. 1B).
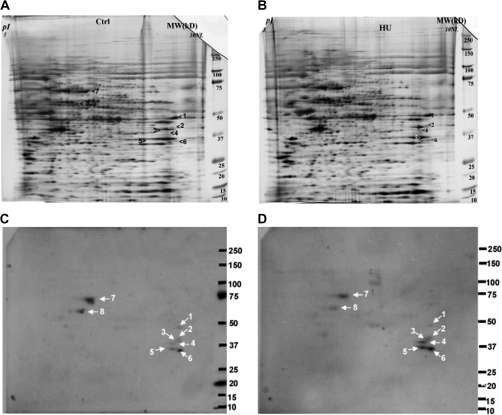
FIG. 2.
2D gel electrophoresis of the tail samples obtained from embryos treated with saline (controls, A) or HU (600 mg/kg, B) and the corresponding 2D Western blots illustrating immunoreactive 4-HNE-protein adducts (C, control; D, HU 600 mg/kg treated). Spots 1–8 were analyzed by MS (see Table 1); (n = 2).
The Identification of 4-HNE–Conjugated Proteins
To identify the 4-HNE-protein adducts enriched in the embryonic tail region, after exposure to vehicle or 600 mg/kg HU, embryo tails were excised and subjected to 2D gel electrophoresis (Fig. 2). 4-HNE–modified protein immunoreactivity revealed eight protein spots in both control and HU-treated embryos. According to the molecular weight range, spots 7 and 8 may be the 4-HNE-protein adducts concentrated in the tail part, whereas spots 1–6 may be more evenly distributed throughout the three embryo regions.
Using MS, identifications were assigned to seven spots (2–8), as summarized in Table 1. The tail-localized 4-HNE–modified proteins were identified as albumin (spot 7), chaperonin subunit theta, and possibly heat shock 60 kDa protein 1 (HSPD1, spot 8). 2D Western blots (n = 2) did not detect a consistent change in the intensity of these three protein adducts when control embryos were compared with HU-exposed embryos. Interestingly, among the lower molecular weight protein adducts (perhaps not tail specific), three of the identified proteins are involved in energy metabolism. These include glutamate oxaloacetate transaminase 2 (GOT2, spot 2); aldolase 1, A isoform (ALDOA1, spot 3); and GAPDH (spot 5). HU exposure resulted in increases in 4-HNE–conjugated GOT2 (1.6-fold), ALDOA1 (1.6-fold), and GAPDH (1.8-fold) compared with controls. The two remaining 4-HNE–modified proteins that were identified are inducible small cytokine subfamily E member 1 (SCYE1, spot 4) and heterogeneous nuclear ribonucleotide protein A1 isoform a (HNRNP A1A, spot 6). HU exposure increased the amounts of both SCYE1 (1.8-fold) and HNRNP A1A (2.1-fold) compared with control.
TABLE 1.
The Identification of Proteins Conjugated with 4-HNE in the Tail Regions of Embryos
| Spot no. | Protein name | Function | MW | pI | Sequence coverage (%) |
| #1 | Unnamed protein | — | 50.4 | 9.1 | 24 |
| #2 | GOT2 | Amino acid metabolism, Kreb's cycle | 47.8 | 9.13 | 28 |
| #3 | ALDOA | Glycolysis | 39.8 | 8.31 | 49 |
| #4 | SCYE1 | Protein translation, apoptosis | 35.5 | 8.75 | 40 |
| #5 | GAPDH | Glycolysis | 36.1 | 8.44 | 44 |
| #6 | HNRNP A1A | RNA processing | 34.3 | 9.27 | 23 |
| #7 | Albumin | Transport protein in serum | 70.7 | 5.75 | 41 |
| #8 | Chaperonin subunit theta | Chaperone | 60.1 | 5.44 | 47 |
| HSPD1 (possible) | Chaperone | 61.1 | 5.91 | 10 |
Note. MW, molecular weight; pI, isoelectric point.
4-HNE Modifications Alter GAPDH Protein
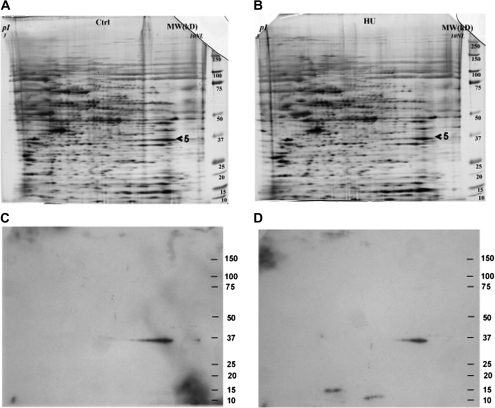
Because glycolysis is a critical energy pathway during organogenesis, we further investigated the importance of GAPDH as a 4-HNE-protein target in the malformation-sensitive tail region of HU-exposed embryos using 2D Western blot analysis (Fig. 3). Western blots of the tail parts of the embryo exposed to saline or HU600 resulted in an immunoreactive spot that matched that of the spot corresponding to the silver-stained gel identified by MS (Figs. 3C and 3D). At 3 h, HU treatment not only increased 4-HNE-protein adducts with GAPDH but also decreased the amount of GAPDH detected and altered the protein conformation of GAPDH, as reflected by a shortened protein migration tail toward the lower isoelectric point.
FIG. 3.
2D gel electrophoresis of the tail samples obtained from embryos treated with saline (control, A) or HU (600 mg/kg, HU600) (B) and the corresponding 2D Western blots illustrating GAPDH immunoreactive protein spots (control, C; HU600, D); (n = 2).
Effects of HU Exposure on GAPDH Enzymatic Activity and Lactate Production
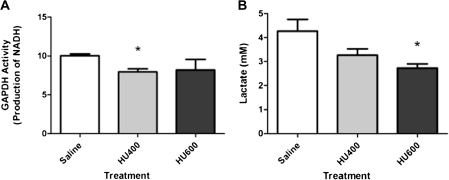
To determine if the formation of GAPDH-4-HNE adducts affected the catalytic activity of GAPDH, we measured its enzymatic activity in the embryo 3 h after exposure of the dams to vehicle or HU. Although GAPDH immunoreactivity in the head, body, and tail were not significantly affected (Supplementary fig. 1), GAPDH activity was significantly decreased in the HU400-treated group (20%), but not the HU600 group, compared with saline (Fig. 4A). The appearance of lactate was measured in whole embryos exposed to HU as a measure of the entire glycolytic pathway because lactate is produced predominantly in anaerobic glycolysis and is an important source of ATP production in the embryo. A 20% decrease in lactate production was observed in embryos exposed to HU400, whereas a 40% decrease occurred in embryos exposed to HU600 (Fig. 4B).
FIG. 4.
(A) Spectrophotometric analysis of NADH as a measure of GAPDH activity in whole embryo samples after treatment with saline (control), low-dose HU (400 mg/kg, HU400), or high-dose HU (600 mg/kg, HU600); (n = 7). (B) Lactate measurements of whole embryo samples treated with saline (control), low-dose HU (400 mg/kg, HU400), or high-dose HU (600 mg/kg, HU600); (n = 3). Asterisk denotes a significant difference between control and treated group (*p < 0.05).
HU Promotes the Nuclear Translocation of GAPDH
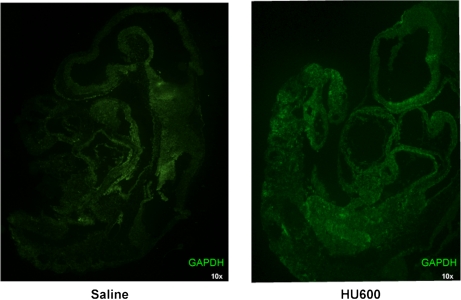
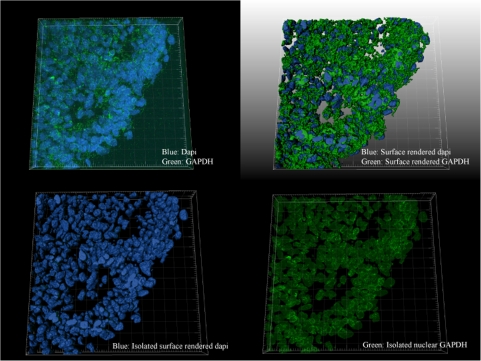
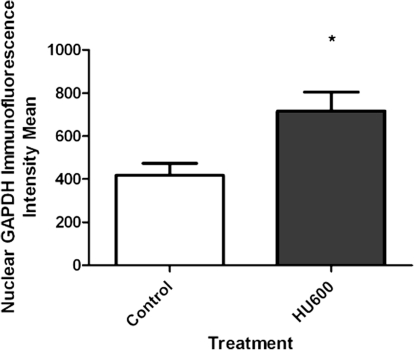
In cell cultures, oxidative stress has been reported to induce the nuclear translocation of GAPDH (Nakajima et al., 2007; Oritz-Oritz et al., 2010); thus, we focused on image analysis to localize GAPDH. Initially, confocal microscopy presented discreet homogeneous spots of immunoreactive GAPDH in both vehicle- and HU-treated embryos, and it was challenging to accurately measure nuclear versus cytoplasmic localization in both treatment groups (Fig. 5). Consequently, we quantified the subcellular localization of GAPDH with z-stack imaging coupled with IMARIS, an advanced 3D imaging software. The lumbosacral somites, the caudal area with the most significant malformations, were the focus of this analysis. A 3D surface render of GAPDH revealed the subcellular localization of GAPDH. In both vehicle- and HU-treated groups, GAPDH immunoreactivity was unevenly distributed within the 3D projection of a given cell; although GAPDH was present both in the cytoplasm and nucleus, its density increased toward the nuclear membrane (Figs. 6A and 6B). A masking technique in IMARIS was applied that subtracted the cytoplasmic from the nuclear staining using DAPI as the nuclear border to quantify nuclear GAPDH (Fig. 6D). Interestingly, although GAPDH was present within the nucleus of nontreated groups, in agreement with the literature, a twofold increase in the nuclear localization of GAPDH was observed in embryos exposed to HU600 compared with saline-treated embryos (Fig. 7).
FIG. 5.
Confocal microscopy images of saline (control, A) or HU (600 mg/kg, HU600, B)-treated embryos. Green fluorescence represents GAPDH immunoreactivity; (n = 5).
FIG. 6.
IMARIS 3D-rendered surface of lumbosacral somites (control sample) to show technique. (top left) Raw data, blue: DAPI, green: GAPDH; (top right) 3D-rendered surface of GAPDH and DAPI combined; (bottom left) 3D-rendered surface of DAPI; (bottom right) 3D surface of DAPI removed leaving nuclear GAPDH; (control n = 6, HU600 n = 7).
FIG. 7.
Intensity mean analysis of nuclear GAPDH, provided by IMARIS software, of embryos treated with saline (control) or HU (600 mg/kg, HU600). Asterisk denotes a significant difference between control and treated group (*p < 0.05).
DISCUSSION
The reactive hydroxylamine group of HU induces ROS, promoting the formation of reactive aldehydes such as 4-HNE through lipid peroxidation (Grimsrud et al., 2008; LoPachin et al., 2009). In previous studies, we reported that 4-HNE-protein adduct immunoreactivity was enhanced in the caudal, malformation-sensitive tissues of embryos exposed to HU. In this study, we identify protein targets of 4-HNE in the tail region of teratogen-exposed embryos. Covalent modifications of 4-HNE to proteins involved in energy metabolism, including GAPDH, were detected.
Interestingly, GAPDH, a protein that is critical in glycolysis in the developing embryo, is susceptible to 4-HNE-protein adduct formation. Furthermore, glycolytic metabolism is reduced in treated embryos. During midorganogenesis (GD 8.5–9.5), the embryo undergoes a metabolic switch from anaerobic to aerobic respiration, coincident with mitochondrial maturation, vascular development, and chorioallantoic coupling (Alcolea et al., 2007; Shepard et al., 1998). At this stage, oxidative phosphorylation has yet to become an important source of energy; indeed, inhibition of the electron transport chain with carbon monoxide on GD 9 does not produce an adverse reaction (Robkin, 1997). Prior to this switch, lactate is the major molecule used in the synthesis of ATP; once this switch has taken place, the reaction equilibrium to pyruvate for transfer into the mitochondria becomes dominant and decreases lactate production. Lactate levels measured in the embryo in this study reflect those of anaerobic glycolysis at GD 9 (4.3 ± 0.8 mmol/l); this is expected to drop (to 2.9 ± 0.3 mmol/l) once the embryo reaches GD 10 (Neubert, 1970). During anaerobic glycolysis, lactate produces only 2 mol of ATP/unit time as compared with aerobic glycolysis, which produces ATP 18× faster. This difference in rate outlines the limitation of anaerobic glycolysis, 2 versus 38 mol of ATP for oxidative phosphorylation. The decrease in glycolytic activity of GAPDH and the subsequent decrease in lactate at this critically susceptible respiratory period suggest that ATP may be deficient during this crucial stage of development. It has been suggested that the reduction of ATP below a critical threshold causes teratogenesis (Ritter et al., 1975).
Another glycolytic target of 4-HNE-protein adducts that has been identified within the caudal region of the embryo is fructose-bisphosphate ALDOA1. There are three ALDO isozymes (A, B, and C); ALDOA1 is mainly produced by the developing embryo and is responsible for the conversion of fructose 1,6-diphosphate into dihydroxyacetone phosphate and glyceraldehyde-3-phosphate in the glycolytic pathway. 4-HNE-protein targets in Parkinson's disease were identified to be ALDOA1 and GAPDH contributing to the metabolic dysfunction of the frontal lobe in the brain (Gómez and Ferrer, 2009); however, there is limited literature reporting that 4-HNE forms protein adducts with ALDOA1. This is the first identification of GAPDH-4-HNE and ALDOA1-4-HNE-protein adducts within the caudal, malformation-sensitive tissue of the developing embryo. Other glycolytic enzymes in muscle, such as enolase, which are a target of 4-HNE-protein adducts, have been shown to have altered enzymatic activity, akin to the enzymatic activity of GAPDH (Hussain et al., 2006). We propose that HU-induced caudal malformations may in part be caused by the aberrant regulation of energy metabolism resulting in cell arrest and death.
In response to the decrease in metabolic function of GAPDH, its nuclear translocation is enhanced. Nuclear translocation of GAPDH is hypothesized to be a stress response during exposure to free radicals and to play a role in the initiation of apoptosis in damaged cells. 2D gel electrophoresis reveals that 4-HNE modifies GAPDH. We hypothesize that GAPDH is primarily modified in the cytoplasm where it comes in direct contact with 4-HNE. Once 4-HNE modifies GAPDH, it is then translocated into the nucleus, whereby it may form new complexes with nuclear proteins, such as p300/CBP and p53, to initiate a cell death signaling cascade. Modifications of GAPDH in the cytoplasm in response to oxidative stress are assumed to be the primary event leading to nuclear localization. 4-HNE–conjugated GAPDH has altered electrophoretic mobility, indicating a conformational change. Accumulating evidence suggests that modifications to GADPH render it an apoptosis executor during oxidative stress (Chuang et al., 2005).
In RAW264.7 cell lines, a murine macrophage-like cell line, GAPDH is S-nitrosylated on cysteine 150, abolishing its enzymatic activity (Sen et al., 2008). Upon S-nitrosylation, GAPDH interacts with Siah1, a protein containing a nuclear localization signal that permits the nuclear translocation of GAPDH because it lacks this sequence. GAPDH becomes acetylated by p300/CBP and mediates apoptosis. The hydroxylamine group of HU has been reported to produce nitric oxide in vivo (Lou et al., 2009); however, whether GAPDH is S-nitrosylated in the fetal embryo at GD 9 and its possible role in mediating the nuclear translocation of GAPDH has yet to be elucidated. In addition to 4-HNE and S-nitrosylation modifications of GAPDH, poly(adenosine diphosphate-ribose) (ADP-ribose) polymerase (PARP) plays an important role in the embryo upon stress. PARP becomes cleaved and activated in response to DNA damage and/or oxidative stress, modifying many specific substrates (Koh et al., 2005). Activated PARP cleaves oxidized nicotinamide adenine dinucleotide (NAD+) to nicotinamide and ADP-ribose and polymerizes the latter on cytoplasmic proteins, nuclear acceptor proteins such as histones, transcription factors, and PARP itself. Poly(ADP-ribosyl)ation contributes to DNA repair and to the maintenance of genomic stability (Wang et al., 1997). However, activation of PARP also depletes cellular NAD+, which subsequently triggers ATP depletion (Ha and Snyder, 1999). Moreover, there is evidence that GAPDH is modified by poly(ADP-ribosyl)ation, causing it to become enzymatically inactivated and leading to further depletion of ATP.
The decrease in energy production in the embryo as a result of the attenuation of glycolysis and the nuclear translocation of GAPDH suggest that GAPDH may serve as an intracellular sensor of oxidation and may play an early and pivotal role in the cascade leading to apoptosis. The translocation of GAPDH from the cytoplasm to the nucleus appears to be a critical step in the induction of apoptosis (Dastoor and Dreyer, 2001). Further insight into protein modifications in region-specific areas of the embryo may better our understanding of its role in oxidative stress–mediated teratogenicity.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Canadian Institutes of Health Research (FRN 57867).
Supplementary Material
Acknowledgments
We thank Leonid Kriazhev, Genome Quebec, for the 2D electrophoresis and mass spectrometric analysis, Ghalib Bardai for the lactate measurements, Cory Glowinski from Bitplane for his diligent efforts and guidance using the IMARIS software, and Anne McKinney from McGill University for her important suggestions and expertise with confocal imaging.
References
- Alcolea MP, Llado I, Garcia-Palmer FJ, Gianotti M. Responses of mitochondrial biogenesis and function to maternal diabetes in rat embryo during the placentation period. Am. J. Physiol. Endocrinol. Metab. 2007;293:E636–E644. doi: 10.1152/ajpendo.00120.2007. [DOI] [PubMed] [Google Scholar]
- Botzen D, Grune T. Degradation of HNE-modified proteins—possible role of ubiquitin. Redox Rep. 2007;12:63–67. doi: 10.1179/135100007X162130. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- Dastoor Z, Dreyer JL. Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J. Cell Sci. 2001;114:1643–1653. doi: 10.1242/jcs.114.9.1643. [DOI] [PubMed] [Google Scholar]
- Dennery PA. Effects of oxidative stress on embryonic development. Birth Defects Res. C Embryo Today. 2007;81:155–162. doi: 10.1002/bdrc.20098. [DOI] [PubMed] [Google Scholar]
- DeSesso JM. Amelioration of teratogenesis. l. Modification of hydroxyurea-induced teratogenesis by the antioxidant propyl gallate. Teratology. 1981;24:19–35. doi: 10.1002/tera.1420240104. [DOI] [PubMed] [Google Scholar]
- DeSesso JM, Scialli AR, Goeringer GC. D-mannitol, a specific hydroxyl free radical scavenger, reduced the developmental toxicity of hydroxyurea in rabbits. Teratology. 1994;49:248–259. doi: 10.1002/tera.1420490404. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Carroll J, Duchen MR, Campbell K, Swann K. Mitochondrial function and redox state in mammalian embryos. Semin. Cell Dev. Biol. 2009;20:346–353. doi: 10.1016/j.semcdb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P. Wnt signaling in limb organogenesis. Organogenesis. 2008;4:109–115. doi: 10.4161/org.4.2.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM. Metabolic reconfiguration is a regulated response to oxidative stress. J. Biol. 2008;7:1. doi: 10.1186/jbiol63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J. Biol. Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez A, Ferrer I. Increased oxidation of certain glycolysis and energy metabolism enzymes in the frontal cortex in Lewy body diseases. J. Neurosci. Res. 2009;87:1002–1013. doi: 10.1002/jnr.21904. [DOI] [PubMed] [Google Scholar]
- Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SN, Matar G, Barreiro E, Florian M, Divangahi M, Vassilakopoulos T. Modifications of proteins by 4-hydroxy-2-nonenal in the ventilatory muscles of rats. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;290:L996–L1003. doi: 10.1152/ajplung.00337.2005. [DOI] [PubMed] [Google Scholar]
- Hwang NR, Yim YM, Jeong J, Song EJ, Lee JH, Choi S, Lee KJ. Oxidative modifications of glyceraldehyde-3-phosphate dehydrogenase play a key role in its multiple cellular functions. Biochem. J. 2009;423:253–264. doi: 10.1042/BJ20090854. [DOI] [PubMed] [Google Scholar]
- Koh DW, Dawson TM, Dawson VL. Poly(ADP-ribosyl)ation regulation of life and death in the nervous system. Cell. Mol. Life Sci. 2005;62:760–768. doi: 10.1007/s00018-004-4508-y. [DOI] [PubMed] [Google Scholar]
- Korswagen HC. Regulation of the Wnt/β-catenin pathway by redox signaling. Dev. Cell. 2006;10:687–688. doi: 10.1016/j.devcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Larouche G, Hales BF. The impact of human superoxide dismutase 1 expression in a mouse model on the embryotoxicity of hydroxyurea. Birth Defects Res. A Clin. Mol. Teratol. 2009;85:800–807. doi: 10.1002/bdra.20595. [DOI] [PubMed] [Google Scholar]
- LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem. Res. Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou TF, Singh M, Mackie A, Li W, Pace BS. Hydroxyurea generates nitric oxide in human erythroid cells: mechanisms for gamma-globin gene activation. Exp. Biol. Med. 2009;234:1374–1382. doi: 10.3181/0811-RM-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Amano W, Fujita A, Azuma YT, Hata F, Inui T, Takeuchi T. The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 2007;282:26562–26574. doi: 10.1074/jbc.M704199200. [DOI] [PubMed] [Google Scholar]
- Neubert D. Aerobic glycolysis in mammalian embryos. In: Bass R, Beck F, Merker HJ, Neubert D, Randhahn B, editors. Metabolic Pathways in Mammalian Embryos during Organogenesis and Its Modifications by Drugs. 1st ed. Berlin, Germany: FU-Berlin; 1970. pp. 225–249. [Google Scholar]
- Novotny MV, Yancey MF, Stuart R, Wiesler D, Peterson RG. Inhibition of glycolytic enzymes by endogenous aldehydes: a possible relation to diabetic neuropathies. Biochim. Biophys. Acta. 1994;1226:145–150. doi: 10.1016/0925-4439(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Oritz-Oritz MA, Moran JM, Ruiz-Mesa LM, Bravo-San Pedro JM, Fuentes JM. Paraquat exposure induces nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the activation of the nitric oxide-GAPDH-Siah cell death cascade. Toxicol. Sci. 2010;116:614–622. doi: 10.1093/toxsci/kfq146. [DOI] [PubMed] [Google Scholar]
- Ozolins TR, Hales BF. Oxidative stress regulates the expression and activity of transcription factor activator protein-1 in rat conceptus. J. Pharmacol. Exp. Ther. 1997;280:1085–1093. [PubMed] [Google Scholar]
- Ritter EJ, Scott WJ, Wilson JG. Inhibition of ATP synthesis associated with 6-aminonicotinamide (6-AN) teratogenesis in rat embryos. Teratology. 1975;12:233–238. doi: 10.1002/tera.1420120304. [DOI] [PubMed] [Google Scholar]
- Robkin MA. Carbon monoxide and the embryo. Int. J. Dev. Biol. 1997;41:283–289. [PubMed] [Google Scholar]
- Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- Schutt F, Bergmann M, Holz FG, Kopitz J. Proteins modified by malondialdehyde, 4-hydroxynonenal, or advanced glycation end products in lipofuscin of human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 2003;44:3663–3668. doi: 10.1167/iovs.03-0172. [DOI] [PubMed] [Google Scholar]
- Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell. Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard TH, Muffley LA, Smith LT. Ultrastructural study of mitochondria and their cristae in embryonic rats and primate (N. nemistrina) Anat. Rec. 1998;252:383–392. doi: 10.1002/(SICI)1097-0185(199811)252:3<383::AID-AR6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Uchida K. Histidine and lysine as targets of oxidative modifications. Amino Acids. 2003;25:249–257. doi: 10.1007/s00726-003-0015-y. [DOI] [PubMed] [Google Scholar]
- Uchida K, Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J. Biol. Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hales BF. Activator protein-1 (AP-1) DNA binding activity is induced by hydroxyurea in organogenesis stage mouse embryos. Toxicol. Sci. 2005;85:1013–1023. doi: 10.1093/toxsci/kfi148. [DOI] [PubMed] [Google Scholar]
- Yan J, Hales BF. Depletion of glutathione induces 4-hydroxynonenal protein adducts and hydroxyurea teratogenicity in the organogenesis stage mouse embryo. J. Pharmacol. Exp. Ther. 2006;319:613–621. doi: 10.1124/jpet.106.109850. [DOI] [PubMed] [Google Scholar]
- Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis. 2000;21:3666–3672. doi: 10.1002/1522-2683(200011)21:17<3666::AID-ELPS3666>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.