Abstract
There is relatively little information regarding the critical xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in Caenorhabditis elegans, despite this organism's increasing use as a model in toxicology and pharmacology. We carried out experiments to elucidate the capacity of C. elegans to metabolically activate important promutagens via CYPs. Phylogenetic comparisons confirmed an earlier report indicating a lack of CYP1 family enzymes in C. elegans. Exposure to aflatoxin B1 (AFB1), which is metabolized in mammals by CYP1, CYP2, and CYP3 family enzymes, resulted in significant DNA damage in C. elegans. However, exposure to benzo[a]pyrene (BaP), which is metabolized in mammals by CYP1 family enzymes only, produced no detectable damage. To further test whether BaP exposure caused DNA damage, the toxicities of AFB1 and BaP were compared in nucleotide excision repair (NER)–deficient (xpa-1) and NER-proficient (N2) strains of C. elegans. Exposure to AFB1 inhibited growth more in xpa-1 than N2 nematodes, but the growth-inhibitory effects of BaP were indistinguishable in the two strains. Finally, a CYP-nicotinamide adenine dinucleotide phosphate reductase–deficient strain (emb-8) of C. elegans was found to be more resistant to the growth-inhibitory effect of AFB1 exposure than N2, confirming that the AFB1-mediated growth inhibition resulted from CYP-mediated metabolism. Together, these results indicate that C. elegans lacks biologically significant CYP1 family–mediated enzymatic metabolism of xenobiotics. Interestingly, we also found that xpa-1 nematodes were slightly more sensitive to chlorpyrifos than were wild type. Our results highlight the importance of considering differences between xenobiotic metabolism in C. elegans and mammals when using this alternative model in pharmaceutical and toxicological research.
Keywords: Caenorhabditis elegans, cytochrome P450, aflatoxin B1, benzo[a]pyrene, genotoxicity, nucleotide excision repair
The nematode Caenorhabditis elegans is emerging as an important model in pharmacology and toxicology (Leung et al., 2008; Peterson et al., 2008). Caenorhabditis elegans is similar to higher eukaryotes in many molecular and cellular pathways (Kaletta and Hengartner, 2006) and offers unique advantages over conventional mammalian models, including the ease of maintenance, short life cycle, genetic manipulability, and high-throughput capability. Caenorhabditis elegans–based assays are increasingly used to evaluate potential toxicity of different stressors in humans (Boyd et al., 2010b; Dengg and van Meel, 2004; Rajini et al., 2008; Sprando et al., 2009) and mechanisms of toxicity after chemical exposures (Cui et al., 2007; Donohoe et al., 2006; Valmas and Ebert, 2006).
A limitation associated with using C. elegans as a model in toxicology is incomplete understanding of its response to human mutagens. The DNA damage response appears to be generally similar in C. elegans and higher eukaryotes (Leung et al., 2008; O'Neil and Rose, 2005; Stergiou and Hengartner, 2004), and some direct-acting DNA-damaging agents that have been commonly used in C. elegans produce comparable responses to those observed in mammals (Ahringer, 2006; Anderson, 1995; Greber et al., 2003; Hartman et al., 1995; Ishiguro et al., 2001; Meyer et al., 2007; Stewart et al., 1991). However, activation-dependent mutagens (i.e., promutagens) have not been well studied in C. elegans and might produce different responses in C. elegans and mammalian models because of differences in xenobiotic metabolism (Lindblom and Dodd, 2006). In particular, Gotoh (1998) provided phylogenetic evidence that C. elegans lacked cytochrome P450 (CYP1) family genes that are responsible for the activation of many promutagens.
Aflatoxin B1 (AFB1) and benzo[a]pyrene (BaP) are two commonly used model promutagens. AFB1 is a naturally occurring mycotoxin found in foods such as corn, peanuts, various other nuts, and cottonseed (Groopman et al., 2005). It remains an important environmental carcinogen in many developing countries (Vineis and Xun, 2009). BaP is a model carcinogenic polycyclic aromatic hydrocarbon (PAH). PAHs are environmental carcinogens that occur at high and increasing levels in the environment and result from incomplete combustion of organic compounds including fossil fuels, wood, cigarettes, and food (Van Metre and Mahler, 2005). AFB1 and BaP share a similar general mechanism of mutagenesis, requiring metabolic activation by CYP enzymes to form epoxide metabolites. The electrophilic epoxides in turn bind to DNA molecules, resulting in bulky, DNA helix–distorting DNA lesions that are repaired by nucleotide excision repair (NER) in the nuclear genome. However, a key difference between AFB1- and BaP-induced DNA damage in mammals is that whereas AFB1 is activated in mammals by CYP1, CYP2, and CYP3 family enzymes, BaP is activated only by CYP1 family enzymes.
Our objective was to investigate the potential role of CYPs in the genotoxicity and metabolism of AFB1 and BaP in C. elegans. We took three complementary approaches. First, we generated a phylogenetic tree of CYPs in C. elegans and other species. Second, we quantified DNA damage caused by exposure to AFB1 and BaP using a quantitative PCR (QPCR)–based assay. Chlorpyrifos (CPF, an organophosphate pesticide) and β-naphthoflavone (BNF, a noncarcinogenic PAH) were also evaluated. Our third approach was to investigate the genotoxicity of AFB1 and BaP exposure in C. elegans using genetic approaches. In the first genetic experiment, we assessed the metabolic activation of AFB1 and BaP in C. elegans in vivo by comparing the relative susceptibility of DNA adduct repair–deficient (xpa-1) and DNA adduct repair–proficient (N2) strains to AFB1 and BaP exposure. In the second genetic experiment, we evaluated the importance of the CYP system in AFB1 activation by comparing the relative susceptibility of CYP-nicotinamide adenine dinucleotide phosphate (NADPH) reductase deficient (emb-8) and wild-type (N2) strains to AFB1 exposure. The results suggested that (1) C. elegans lacks CYP1 family enzymes; (2) AFB1, but not BaP, produced a biologically significant level of DNA adducts; and (3) the CYP system played an important role in activating AFB1 in C. elegans. This important difference between the xenobiotic metabolism of C. elegans and higher eukaryotes needs to be taken into account when using this alternative model in pharmaceutical and toxicological research.
MATERIALS AND METHODS
Phylogenetic analysis.
Gene models in publicly available nematode genomes were searched using Hmmer (v2.3.2: Eddy, 1998). Amino acid sequences were aligned using Muscle (v3.6: Edgar, 2004) and automatically masked based on the alignment quality score assigned by Muscle. A maximum likelihood phylogenetic tree was constructed with RAxML using the Whelan and Goldman model of amino acid substitution and a gamma distribution of rate categories (Stamatakis, 2006). Previously unnamed nematode CYPs in C. briggsae were assigned names by the Cytochrome P450 Nomenclature Committee and are available at the Cytochrome P450 homepage (Nelson, 2009); CYPs in Meloidogyne incognita, and Brugia malayi have not been formally named yet.
Caenorhabditis elegans culture.
The wild-type N2 (Bristol), emb-8 (CYP-NADPH reductase–deficient MJ69), and glp-1 (germ line–deficient JK1107) strains of C. elegans were obtained from the Caenorhabditis Genetics Center (University of Minnesota). xpa-1 (NER-deficient strain RB864) was previously outcrossed three times (Meyer et al., 2007). Populations of C. elegans were maintained on K agar plates seeded with OP50 bacteria (Lewis and Fleming, 1995) at 20°C unless otherwise stated. Semisynchronized populations of nematodes were obtained by bleach-sodium hydroxide isolation of eggs (Lewis and Fleming, 1995). L1 growth-arrested (starved) larvae were obtained by hatching eggs in complete K-medium (Boyd et al., 2009) overnight with shaking (Lewis and Fleming, 1995). All transfers were made by washing nematodes off of agar plates and rinsing in K-medium (Williams and Dusenbery, 1990) after centrifugation at 2000 × g for 2 min.
Chemical exposures.
AFB1, BaP, CPF, and BNF (Sigma Chemical Co., St Louis, MO) were dissolved in dimethyl sulfoxide (DMSO) to prepare stock solutions. Three hundred glp-1 adults were dispensed into each well of a 12-well plate. Each well contained a mixture of 990 μl complete K-medium, 10 μl stock solution dissolved in DMSO, and OP50. The 1% DMSO was found not to affect nematode growth or reproduction (data not shown). The exposure concentrations were selected based on preliminary lethality assays (data not shown) or solubility, such that the highest concentration was either that which first showed mortality or the highest achievable based on solubility if lethality could not be reached. This was the case for AFB1 and BaP, which had solubility limits of ∼100μM in complete K-medium with 1% DMSO. Caenorhabditis elegans showed normal behavior at all concentrations of AFB1, BaP, and BNF and lower concentrations of CPF but were paralyzed at 100μM of CPF.
QPCR-based DNA damage assay.
Nuclear DNA damage was evaluated using a QPCR-based method (Meyer et al., 2007) as adapted for use in a small number of individual nematodes (Boyd et al., 2010a; Hunter et al., 2010). This assay defines the control samples as undamaged and generates a lesion frequency in experimental samples based on a decrease in amplification efficiency relative to the control samples and has previously been used to detect BaP-induced DNA damage (Jung et al., 2009a,b). Two nuclear genome targets (unc-2 and small nuclear, 9316 and 225 nt, respectively; Meyer et al., 2007) were amplified. The amount of long PCR product provides a measurement of lesion frequency, whereas the amount of short PCR product provides normalization to DNA template amount. Lesion calculations were performed as described previously (Ayala-Torres et al., 2000; Meyer, 2010). Nematodes were sampled after 48-h exposures. These experiments were carried out using a temperature-sensitive mutant strain (glp-1) in which maintenance at 25°C blocks germ line proliferation and therefore blocks cell division because outside of the germ line, no cell divisions occur in adult C. elegans (Sulston, 1988). Because young adult C. elegans have a rapidly proliferating germ line, DNA damage caused by chemical exposure could be readily “diluted” by the new DNA produced by dividing germ cells, confounding measurements of DNA damage (Meyer et al., 2007). Six adults were pooled for each biological replicate, and four biological replicates were taken per treatment. A total of eight biological replicates per treatment were used in the analysis.
Growth assay.
Two genetic experiments were carried out to investigate (1) the effects of AFB1, BaP, CPF, and BNF on NER-deficient (xpa-1) and NER-proficient (N2) strains of C. elegans and (2) the effect of AFB1 on CYP-NADPH reductase–deficient (emb-8) and wild-type (N2) strains of C. elegans. The growth of C. elegans was assessed essentially as previously described (Smith et al., 2009). In both experiments, growth inhibition was measured as an indicator of chemical-induced genotoxicity because xpa-1 larval growth is dramatically impaired by DNA damage that requires NER proteins for removal (Astin et al., 2008).
In the first growth assay, L1 N2 and xpa-1 nematodes were transferred to the sample cup of the COPAS Biosort (Union Biometrica Inc., Somerville, MA) and diluted to approximately 1 nematode per microliter. Fifty L1s were then added to each well of a 96-well plate containing a total volume of 50 μl complete K-medium, OP50, and chemical stock solution. Caenorhabditis elegans cohorts were incubated for 48 h at 20°C, and then size measurements of individual nematodes were acquired with the COPAS Biosort ReFLEx as previously described (Boyd et al., 2009).
The second growth assay was conducted using L1 N2 and emb-8 nematodes. The nematodes were hatched overnight at 15°C and then transferred to unseeded 100 mm K agar plates containing solvent control (1% vol/vol DMSO), 30μM AFB1, and 100μM AFB1 and incubated at 23°C for 2 days. The MJ69 strain carries a temperature-sensitive mutation in the emb-8 gene such that the phenotype is essentially normal at 15°C, but CYP-NADPH reductase activity is impaired at and above 23°C (Kulas et al., 2008). The animals were then transferred to seeded K agar plates, incubated at 15°C for 2 days, and photographed using a Nikon Eclipse E600 camera (Tokyo, Japan). The length of the nematode was determined using Lucia 5 (Laboratory Imaging, Prague, Czech). Two separate experiments were conducted and the results combined.
Statistical analysis.
All data were analyzed with Statview for Windows (Version 5.0.1, SAS Institute Inc., Cary, NC). DNA damage data were assessed using an initial two- or three-way ANOVA (ANOVA on exposure level and time point, as well as presence/absence of bacteria in the case of the AFB1 exposure) with a Bonferroni correction for five multiple comparisons (four chemicals plus presence/absence of bacteria for AFB1). Post hoc analysis was carried out using Fisher's Protected Least Significant Differences test. Growth data were not normally distributed (as assessed by the Kolmogorov-Smirnov Normality test) and so were analyzed using Mann-Whitney U or Kruskal-Wallis tests followed by Bonferroni corrections for multiple comparisons. p Values < 0.05 (after Bonferroni corrections) were considered significant. Box plots indicate 10th, 25th, 50th, 75th, and 90th percentiles, plus outliers.
RESULTS
Lack of Gene Sequence–Based Evidence for CYP1 Family CYPs in C. elegans
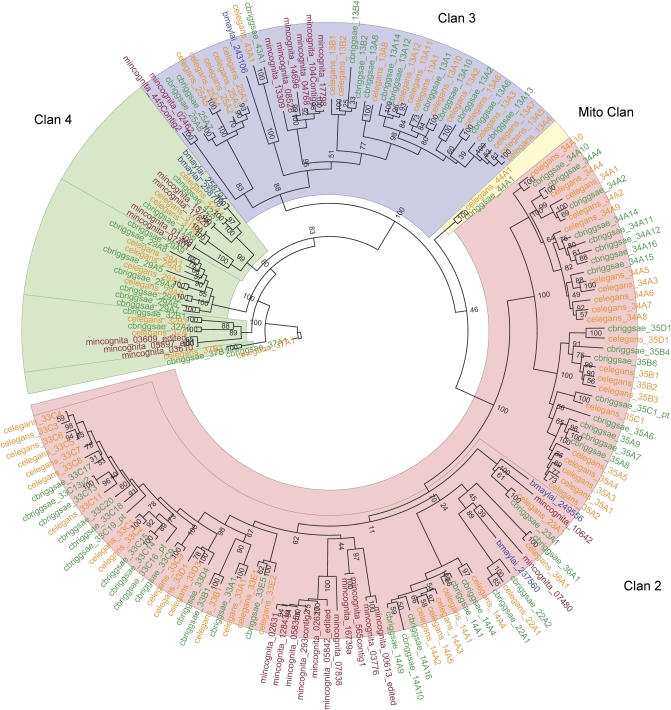
Previous investigations have found no evidence for CYP1 family genes in nonchordates (Goldstone et al., 2007). Our investigation of the CYP complements of the four nematode genomes reported here (C. elegans, C. briggsae, M. incognita, and B. malayi) support the fact that CYP1s are not present in the nematode genomes. A phylogenetic tree of the CYP complements of the four nematodes demonstrates that CYP1 genes are not present, although a large number of CYP2-like (Clan 2) genes are present and expressed in C. elegans (Fig 1). Many CYP2 genes in vertebrates are xenobiotic (drug)-metabolizing genes, and at least one (CYP2S1) is inducible via the important xenobiotic-responsive transcription factor aryl hydrocarbon receptor (AHR; Saarikoski et al., 2005).
FIG. 1.
Maximum likelihood phylogeny of CYPs from four nematode genomes, including the free-living C. elegans (orange) and C. briggsae (green) and the parasitic Meloidogyne incognita (red) and Brugia malayi (blue). The CYP Clan 2 genes, related to vertebrate xenobiotic-metabolizing CYP2s, are highlighted in yellow. Values at node points are bootstrap values (100 replicate bootstraps, randomly seeded).
AFB1 Exposure Results in DNA Damage
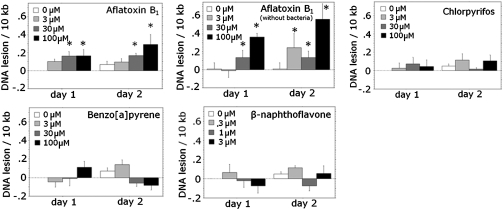
To empirically test the prediction of our phylogenetic analysis, we measured DNA damage after exposure to promutagens requiring (BaP) and not requiring (AFB1) CYP1-like activity for activation using a QPCR assay (Hunter et al., 2010). This assay detects any DNA lesions that significantly inhibit the progression of the DNA polymerase used in the PCR reaction. AFB1 exposure resulted in concentration-dependent DNA damage (p = 0.0007 for main effect of concentration, two-factor ANOVA) in C. elegans. Damage was detectable after exposures of 30 and 100μM AFB1. BaP, BNF, and CPF exposure did not result in any detectable DNA damage (p = 0.615, 0.161, and 0.454, respectively, for the effect of concentration) (Fig 2). The limit of detection of the QPCR assay is approximately 1 lesion per 105 bases (Hunter et al., 2010).
FIG. 2.
DNA damage is caused by exposure to AFB1 (with and without bacteria), but not BaP, BNF, or CPF in C. elegans. Young adult glp-1 nematodes were exposed for 48 h in liquid medium and sampled at 24 and 48 h (total n = 8 nematodes per concentration per chemical per time point). AFB1 exposure in C. elegans resulted in concentration-dependent DNA damage (p < 0.001, main effect of concentration in two-factor ANOVA); concentrations at which the AFB1-induced DNA damage measured was significantly different from controls (p < 0.05 by Fisher's Protected Least Significant Differences) are indicated by asterisks. BaP, BNF, and CPF exposure did not result in a detectable level of DNA damage (p = 0.615, 0.161, and 0.454 respectively). The experiment was carried out twice (n = 4 each) and the results combined.
In order to determine whether the OP50 strain of Escherichia coli (i.e., the C. elegans food source) might be responsible for the production of carcinogenic AFB1 metabolites in our experimental system, we repeated AFB1 exposure without adding bacteria to the exposure medium (Fig. 2). The exclusion of bacteria did not abrogate the induction of DNA damage (p = 0.0005 for main effect of concentration, two-factor ANOVA on OP50-fed nematodes only), indicating that C. elegans was responsible for metabolizing AFB1 to the activated form. In fact, exposure without bacteria actually resulted in a slightly greater level of DNA damage than exposure with bacteria (p = 0.039 for interaction of presence of bacteria and concentration, three-factor ANOVA).
DNA Repair–Deficient Nematodes Are More Sensitive than Wild Type to the Growth-Inhibitory Effects of AFB1 and CPF but Not BaP or BNF
It remained possible that BaP, BNF, or CPF caused DNA damage at a level not detected by QPCR but nonetheless biologically relevant. To test this possibility, we employed the xpa-1 strain. The xpa-1 strain carries a large deletion in the nematode homolog of the xeroderma pigmentosum group A gene, which is required for NER (Berneburg and Lehmann, 2001). Many structurally dissimilar environmental genotoxins, including PAHs such as BaP, mycotoxins such as AFB1, and ultraviolet C radiation can produce helix-distorting DNA lesions that are removed by NER (Hanawalt, 2002; Sancar and Reardon, 2004). xpa-1 nematodes are exquisitely sensitive to DNA damage that is repaired by the NER pathway (Astin et al., 2008; Boyd et al., 2010a; Hartman and Herman, 1982; Meyer et al., 2007). In particular, larval growth of xpa-1 nematodes is highly sensitive to such DNA damage (Astin et al., 2008). Therefore, if any of these chemicals cause biologically significant helix-distorting DNA damage, xpa-1 nematodes would show more growth inhibition than N2.
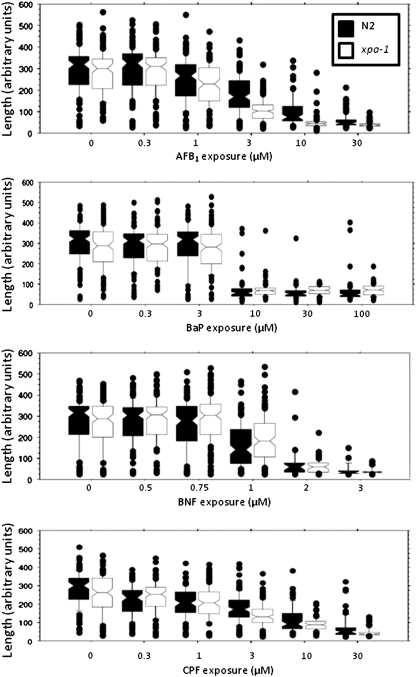
Exposure levels of AFB1, BaP, BNF, and CPF that would lead to larval growth inhibition in the wild-type N2 strain were identified first. BNF caused the strongest growth-inhibitory effects (Fig 3 and Supplementary fig. 2), causing a > 40% size reduction as compared with controls at the concentration of 1μM (based on comparison of median values). The length of nematodes as measured by time of flight is shown in Figure 3; their optical density (extinction) is shown in Supplementary figure 2, and detailed statistical information is presented in Supplementary table 1. Exposures to AFB1, CPF, and BaP resulted in a similar growth-inhibitory effect at the concentrations of 3, 3, and 10μM, respectively.
FIG. 3.
AFB1 and CPF inhibited growth more in a DNA repair–deficient strain (xpa-1, white) than in the wild-type (N2, black) strain of C. elegans. Exposure to BaP and BNF inhibited growth of both strains to a statistically indistinguishable degree; n = 25–143 nematodes per concentration per strain per chemical; results include three separate (pooled) experiments. See Supplementary table 1 for statistical details. Size measurements were taken on day 2 after feeding began and are presented here as length (time of flight) measurements. For optical density–based growth measurements, see Supplementary figure 2.
As shown in Figure 3, exposure to AFB1 and CPF resulted in a greater growth inhibition in xpa-1 as compared with N2. BaP and BNF resulted in comparable responses in N2 and xpa-1 (p > 0.05 for N2 vs. xpa-1 at all concentrations for all three chemicals). Because larval growth inhibition is a very sensitive indicator of DNA damage in xpa-1 nematodes and sensitivity to DNA damage is the only phenotype documented in xpa-1 nematodes (Boyd et al., 2010a), these results suggest that AFB1 and CPF, but not BaP or BNF, produced DNA damage (of the type repaired by NER) at a biologically significant level in C. elegans.
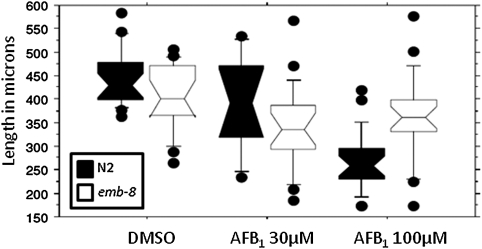
AFB1-Mediated Larval Growth Inhibition Is Partially Rescued in Nematodes Deficient in CYP-NADPH Reductase Activity
We hypothesized that AFB1 activation to a genotoxic form was CYP mediated based on the presence of CYP2 and CYP3 family homologs in C. elegans. To test this hypothesis directly, we compared the effect of AFB1 toxicity in N2 and emb-8 nematodes. emb-8 nematodes carry a point mutation in the gene coding for CYP-NADPH reductase (Rappleye et al., 2003), resulting in temperature-sensitive disruption of function. Because AFB1 activation via CYP catalytic activity requires CYP-NADPH reductase, emb-8 mutants are deficient in CYP activity at the nonpermissive temperature (Kulas et al., 2008). Exposure to AFB1 resulted in less growth inhibition in the emb-8 than the N2 strain (Fig 4), confirming a role for CYP enzymes in AFB1 toxicity. AFB1 inhibited growth in both strains (p < 0.0001 and p = 0.0006 for N2 and emb-8, respectively, Kruskal-Wallis test). However, although emb-8 nematodes were somewhat smaller than N2 under control conditions (emb-8 median ∼86% of N2; p = 0.0002, Mann-Whitney U-test), they were larger after exposure to 100μM AFB1 (emb-8 median ∼140% of N2; p = 0.0007). There was no difference in size at 30μM AFB1 (p = 0.1376).
FIG. 4.
AFB1 inhibited the growth of a CYP-NADPH reductase–deficient strain (emb-8, white) less effectively than growth of wild-type (N2, black) C. elegans (p = 0.0002, 0.1376, and 0.0007, strain comparisons at 0, 30, and 100μM AFB1 by Mann-Whitney U-test); n = 17–24 nematodes, two separate biological experiments pooled.
DISCUSSION
Caenorhabditis elegans Appears to Lack CYP1 Family Enzymes and the Corresponding Ability to Enzymatically Activate the Procarcinogen BaP
CYPs play critical roles in normal metabolism as well as in xenobiotic metabolism. Our phylogenetic analysis suggests that although C. elegans has a large number of CYPs (83), it lacks family 1 genes. Our molecular and genetic experiments indicated that BaP, an environmentally important and well-studied promutagenic PAH, is not activated to a DNA-reactive form at biologically significant rates in C. elegans, indicating that C. elegans lacks a CYP capable of this CYP1-like activity.
A previous study by Gotoh (1998) also failed to identify CYP1 family homologs in C. elegans. However, Chakrapani et al. (2008) suggested that C. elegans contains a CYP1A2 homolog and found that this gene (cyp-14A3) was induced by both BaP and (to a lesser extent) BNF. In addition, Schäfer et al. (2009) showed that cyp-14A3 and related genes were able to hydroxylate polychlorinated biphenyl (PCB52). Finally, improved and much-expanded sequence data have become available for C. elegans and other nematode and nonnematode species. Therefore, we carried out additional phylogenetic analyses but still failed to identify any CYP1 family genes in C. elegans. Nematodes have other Clan 2 genes, including the CYP2-like CYP14, CYP33, CYP34, and CYP35 families (Abad et al., 2008; Gotoh, 1998). In particular, C. elegans CYP35 genes are responsive to a variety of xenobiotic stressors (Menzel et al., 2001, 2005; Reichert and Menzel, 2005), and a number of other CYPs have been shown via microarray to be induced by PCB52 (Menzel et al., 2007), including members of families CYP13, CYP14, CYP25, CYP29, CYP33, CYP34, and CYP37.
The Promutagen AFB1 Causes DNA Damage Detectable by QPCR Analysis in C. elegans, but BaP Does Not
AFB1 and BaP are both promutagens that require metabolic activation before reacting with DNA. AFB1 and BaP are similar in size and structure, both requiring addition of an epoxy group to become DNA reactive (Supplementary fig. 1). The electrophilic epoxy metabolites attack the nucleophilic centers of the DNA molecule, such as the ring nitrogen (i.e., N7) of guanine. The resultant large DNA adducts, often referred to as “bulky lesions,” distort the DNA helix and can interfere with DNA transcription and replication. Some can also detach along with the adducted base from the DNA strand, resulting in abasic sites. Although the metabolic activation of both AFB1 and BaP in mammals requires CYP-mediated hydroxylation, different CYP family members are involved. The activation of AFB1, for instance, can be carried out by mammalian CYP1A2, CYP2A6, CYP2B6, and CYP3A4 (Egner et al., 2003; Mace et al., 1997). In contrast, the activation of BaP (and other PAHs) in mammals is mainly catalyzed by CYP1 family enzymes, especially CYP1B1 and CYP1A1 (Shimada, 2006; Shimada and Fujii-Kuriyama, 2004).
Our results indicate that C. elegans can metabolize AFB1 into DNA-binding metabolites and that this activation is CYP dependent. We have previously observed that xpa-1 nematodes are more sensitive than N2 to AFB1-induced growth inhibition (Meyer et al., 2010) and here extend that result with more extensive growth analysis, direct measurements of DNA damage, and genetic data indicating that the AFB1 activation is CYP mediated. In contrast, C. elegans cannot activate BaP, at least not sufficiently to lead to DNA damage detectable by the QPCR assay. Although it is impossible to entirely rule out the possibility that some low amount of BaP-metabolizing capacity exists in C. elegans, the lack of a growth-inhibitory effect in the xpa-1 strain indicates that any such capacity that might exist is too small to be biologically relevant for C. elegans. A similar apparent lack of effect of BaP was previously observed by Miller and Hartman, (1998) working with the independently isolated rad-3 (allelic to xpa-1: Astin et al., 2008) strain, as well as with additional radiation-sensitive strains of C. elegans.
Because BNF is not a carcinogenic PAH, it was not surprising that BNF exposure resulted in no detectable DNA damage or differential inhibition of growth in xpa-1 nematodes. We did not detect statistically significant DNA damage after CPF exposure by QPCR analysis, but the xpa-1 nematodes were somewhat more sensitive than wild type to CPF-induced growth inhibition (although the difference was quantitatively less than for AFB1). There is evidence that exposure to CPF may result in oxidative DNA damage under some circumstances (Crumpton et al., 2000); our results support the likelihood that high concentrations of CPF (close to those that caused paralysis in our experiments) can cause DNA damage. It is unclear why xpa-1 growth was more inhibited than N2 growth by CPF, despite a lack of detectable DNA damage as assessed by QPCR. We have previously shown that xpa-1 nematodes have very few if any phenotypes in unstressed conditions, yet are highly sensitive to DNA damage (Boyd et al., 2010a). It is conceivable, however, that there is a phenotype that can only be observed after exposure to a neurotoxin. Neurodegeneration is one of relatively few phenotypes observed in NER-deficient humans, and there is evidence that this may result at least in part from unusual types of oxidative DNA damage that are only repaired by NER (Brooks, 2008). Other potential explanations for the discrepancy would be if the growth assay is more sensitive than the QPCR assay or if CPF causes a type of DNA damage that the QPCR assay detects inefficiently (Meyer, 2010).
Comparative Biology of CYP1 Family Activity and PAH Metabolism in C. elegans
Some invertebrates do metabolize common vertebrate CYP1 family substrates such as BaP, although typically relatively slowly compared with vertebrates (den Besten, 1998; Jorgensen et al., 2005; Little et al., 1985; McElroy, 1990); many others do not (James and Boyle, 1998; Lee, 1998; Rewitz et al., 2006). Although BNF and BaP were both shown to induce some CYPs in C. elegans (Menzel et al., 2001), C. elegans would appear to be among the invertebrates that do not metabolize BaP. Another important difference between C. elegans (and many other invertebrates) and higher eukaryotes is that C. elegans homologs of the AHR do not bind to 2,3,7,8-tetrachlorodibenzo-p-dioxin or BNF (Butler et al., 2001; Powell-Coffman et al., 1998). Thus, the CYP induction and growth inhibition resulting from these two chemicals are presumably AHR independent. The physiological significance of the AHR pathway in C. elegans is currently relatively poorly understood, although there is evidence that it plays a role in developmental neurobiology (Huang et al., 2004; Qin and Powell-Coffman, 2004; Qin et al., 2006). Similarly, the gene regulatory pathways controlling CYP expression in C. elegans will be an important area of future research both from the perspective of using C. elegans as a model organism and to understand the evolution and function of the C. elegans response to environmental cues (Braendle et al., 2008).
Toxicity of AFB1, BaP, CPF, and BNF in C. elegans
BNF was the most potent growth inhibitor in our study and BaP the least. That finding appears to contradict the observation of Menzel et al., (2001) in which the effective concentration (EC)10 values of BaP and BNF in a reproductive assay were 1 and 18μM, respectively. We carried out preliminary studies to test the effect of AFB1, BaP, CPF, and BNF on reproduction using published methods (Boyd et al., 2010b) and found a similar order of reproductive toxicity as for growth inhibition (BNF > AFB1 ≈ CPF > BaP, with xpa-1 more sensitive than N2 only to AFB1). Therefore, the difference between our rank order and that of Menzel et al. (2001) presumably results from differences in experimental procedures.
Although BaP exposure did not result in detectable DNA adducts in C. elegans, it did inhibit the growth of C. elegans. This likely occurred via a nongenotoxic mechanism because xpa-1 nematodes were no more sensitive than wild type. One possibility is that BaP caused narcosis (Di Toro et al., 2000; Schultz, 1989), although we do not have data to indicate either how much BaP is taken up by C. elegans or at what level BaP causes narcosis in this species. The presumably very slow metabolism of BaP in C. elegans increases the likelihood of this possibility. Another possibility is altered gene expression. Menzel et al. (2001), for instance, reported that BaP can induce CYP35 expression in C. elegans at 1μM. Although the functional consequences of CYP35 (and other gene) induction require further investigation, it is possible that it may interfere with developmental processes in C. elegans; PAHs are potent developmental toxicants in some species, and not all act via AHR agonism (Billiard et al., 2008).
Similarly, the mechanism of toxicity of BNF in C. elegans is unclear because it presumably does not act via AHR agonism, the best described mode of action of this chemical. Like BaP, it may also act through altered gene transcription. It affects expression of CYPs and many other genes in C. elegans and other invertebrates (Reichert and Menzel, 2005; Watanabe et al., 2008).
Implications and Conclusions
We identified an important difference in chemical mutagenesis between the model organism C. elegans and vertebrates, resulting from differences in CYP-mediated xenobiotic metabolism. Although both AFB1 and BaP are routinely used in mammalian models in cancer research, exposure to AFB1 but not BaP resulted in detectable DNA damage through metabolic activation in C. elegans. Our results suggest that CYP1 family–like enzymatic activities in general are lacking in C. elegans. If so, this will result in altered pharmacokinetics and toxicokinetics for many important xenobiotics, causing either more or less toxicity as compared with most vertebrates because of decreased clearance and/or decreased metabolic activation. This finding highlights the importance of considering xenobiotic metabolism in the interpretation of toxicological data from this alternative model.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (R21 NS065468 to J.N.M.); National Toxicology Program (Z01ES102046 to W.A.B.); Intramural Research Program of the National Institute of Environmental Health Sciences (Z01ES102045 to J.H.F.); National Institutes of Health Grants to John Stegeman (R01-ES015912, Superfund Basic Research Program at Boston University 5-P42-ES007381) to J.V.G.; Caenorhabditis Genetics Center, National Center for Research Resources.
Acknowledgments
We thank Lauren P. Battle, Avery M. Berkowitz, Bryan W. Clark, Cole W. Matson, Madeline G. McKeever, Julie R. Rice, Elizabeth C. Shamsheldin, Marjolein V. Smith, and Xinyu Yang for their advice and assistance in this study.
References
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Ahringer J. Reverse genetics. In: Community TCER, editor. WormBook. 2006. doi:/10.1895/wormbook.1891.1847.1891. [Google Scholar]
- Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. [PubMed] [Google Scholar]
- Astin JW, O'Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair (Amst.) 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- Billiard SM, Meyer JN, Wassenberg DM, Hodson PV, Di Giulio RT. Non-additive effects of PAHs on early vertebrate development: mechanisms and implications for risk assessment. Toxicol. Sci. 2008;105:5–23. doi: 10.1093/toxsci/kfm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Lehmann DW, Leung MC-K, Rodriguez A, Freedman JH, Van Houten B, Meyer JN. Nucleotide excision repair is not detectably inducible, but is required for normal lifespan and growth, in genotoxin-stressed adult Caenorhabditis elegans. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2010a;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, Rice JR, Snyder DW, Freedman JH. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 2010b;245:153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. Application of a mathematical model to describe the effects of chlorpyrifos on Caenorhabditis elegans development. PLoS ONE. 2009;4:e7024. doi: 10.1371/journal.pone.0007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C, Milloz J, Felix MA. Mechanisms and evolution of environmental responses in Caenorhabditis elegans. Curr. Top. Dev. Biol. 2008;80:171–207. doi: 10.1016/S0070-2153(07)80005-6. [DOI] [PubMed] [Google Scholar]
- Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst.) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler RA, Kelley ML, Powell WH, Hahn ME, Van Beneden RJ. An aryl hydrocarbon receptor (AHR) homologue from the soft-shell clam, Mya arenaria: evidence that invertebrate AHR homologues lack 2,3,7,8-tetrachlorodibenzo-p-dioxin and beta-naphthoflavone binding. Gene. 2001;278:223–234. doi: 10.1016/s0378-1119(01)00724-7. [DOI] [PubMed] [Google Scholar]
- Chakrapani BP, Kumar S, Subramaniam JR. Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J. Biosci. 2008;33:269–277. doi: 10.1007/s12038-008-0044-5. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Is oxidative stress involved in the developmental neurotoxicity of chlorpyrifos? Dev. Brain Res. 2000;121:189–195. doi: 10.1016/s0165-3806(00)00045-6. [DOI] [PubMed] [Google Scholar]
- Cui YX, McBride SJ, Boyd WA, Alper S, Freedman JH. Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 2007;8:R122. doi: 10.1186/gb-2007-8-6-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten PJ. Cytochrome P450 monooxygenase system in echinoderms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 1998;121:139–146. doi: 10.1016/s0742-8413(98)10034-8. [DOI] [PubMed] [Google Scholar]
- Dengg M, van Meel JCA. Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. J. Pharmacol. Toxicol. 2004;50:209–214. doi: 10.1016/j.vascn.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Di Toro DM, McGrath JA, Hansen DJ. Technical basis for narcotic chemicals and polycyclic aromatic hydrocarbon criteria. I. Water and tissue. Environ. Toxicol. Chem. 2000;19:1951–1970. [Google Scholar]
- Donohoe DR, Aamodt EJ, Osborn E, Dwyer DS. Antipsychotic drugs disrupt normal development in Caenorhabditis elegans via additional mechanisms besides dopamine and serotonin receptors. Pharmacol. Res. 2006;54:361–372. doi: 10.1016/j.phrs.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner PA, Yu X, Johnson JK, Nathasingh CK, Groopman JD, Kensler TW, Roebuck BD. Identification of aflatoxin M1-N7-guanine in liver and urine of tree shrews and rats following administration of aflatoxin B1. Chem. Res. Toxicol. 2003;16:1174–1180. doi: 10.1021/tx034106u. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Goldstone HMH, Morrison AM, Tarrant A, Kern SE, Woodin BR, Stegeman JJ. Cytochrome p450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol. Biol. Evol. 2007;24:2619–2631. doi: 10.1093/molbev/msm200. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Divergent structures of Caenorhabditis elegans cytochrome P450 genes suggest the frequent loss and gain of introns during the evolution of nematodes. Mol. Biol. Evol. 1998;15:1447–1459. doi: 10.1093/oxfordjournals.molbev.a025872. [DOI] [PubMed] [Google Scholar]
- Greber B, Lehrach H, Himmelbauer H. Characterization of trimethylpsoralen as a mutagen for mouse embryonic stem cells. Mutat. Res. 2003;525:67–76. doi: 10.1016/s0027-5107(02)00316-0. [DOI] [PubMed] [Google Scholar]
- Groopman JD, Johnson D, Kensler TW. Aflatoxin and hepatitis B virus biomarkers: a paradigm for complex environmental exposures and cancer risk. Cancer Biomark. 2005;1:5–14. doi: 10.3233/cbm-2005-1103. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Dewilde D, Dwarakanath VN. Genetic and molecular analyses of UV radiation-induced mutations in the fem-3 gene of Caenorhabditis elegans. Photochem. Photobiol. 1995;61:607–614. doi: 10.1111/j.1751-1097.1995.tb09876.x. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Herman RK. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics. 1982;102:159–178. doi: 10.1093/genetics/102.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Powell-Coffman JA, Jin Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development. 2004;131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Hunter S, Jung D, Di Giulio R, Meyer J. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Yasuda K, Ishii N, Ihara K, Ohkubo T, Hiyoshi M, Ono K, Senoo-Matsuda N, Shinohara O, Yosshii F, et al. Enhancement of oxidative damage to cultured cells and Caenorhabditis elegans by mitochondrial electron transport inhibitors. IUBMB Life. 2001;51:263–268. doi: 10.1080/152165401753311816. [DOI] [PubMed] [Google Scholar]
- James MO, Boyle SM. Cytochromes P450 in crustacea. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998;121:157–172. doi: 10.1016/s0742-8413(98)10036-1. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, Giessing AMB, Rasmussen LJ, Andersen O. Biotransformation of the polycyclic aromatic hydrocarbon pyrene in the marine polychaete Nereis virens. Environ. Toxicol. Chem. 2005;24:2796–2805. doi: 10.1897/05-047r.1. [DOI] [PubMed] [Google Scholar]
- Jung D, Cho Y, Collins LB, Swenberg JA, Di Giulio RT. Effects of benzo[a]pyrene on mitochondrial and nuclear DNA damage in Atlantic killifish (Fundulus heteroclitus) from a creosote-contaminated and reference site. Aquat. Toxicol. 2009a;95:44–51. doi: 10.1016/j.aquatox.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Cho Y, Meyer JN, Di Giulio RT. The long amplicon quantitative PCR for DNA damage assay as a sensitive method of assessing DNA damage in the environmental model, Atlantic killifish (Fundulus heteroclitus) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009b;149:182–186. doi: 10.1016/j.cbpc.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T, Hengartner MO. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- Kulas J, Schmidt C, Rothe M, Schunck WH, Menzel R. Cytochrome P450-dependent metabolism of eicosapentaenoic acid in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2008;472:65–75. doi: 10.1016/j.abb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Lee RF. Annelid cytochrome P-450. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 1998;121:173–179. doi: 10.1016/s0742-8413(98)10037-3. [DOI] [PubMed] [Google Scholar]
- Leung MC-K, Williams PL, Benedetto A, Au C, Helmke KJ, Aschner M, Meyer JN. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic culture methods. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: pp. 3–29. Academic Press; 1995. [Google Scholar]
- Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. A Comp. Exp. Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little PJ, James MO, Pritchard JB, Bend JR. Temperature-dependent disposition of [14C]benzo(a)pyrene in the spiny lobster, Panulirus argus. Toxicol. Appl. Pharmacol. 1985;77:325–333. doi: 10.1016/0041-008x(85)90332-1. [DOI] [PubMed] [Google Scholar]
- Mace K, Aguilar F, Wang JS, Vautravers P, Gomez-Lechon M, Gonzalez FJ, Groopman J, Harris CC, Pfeifer AM. Aflatoxin B1-induced DNA adduct formation and p53 mutations in CYP450-expressing human liver cell lines. Carcinogenesis. 1997;18:1291–1297. doi: 10.1093/carcin/18.7.1291. [DOI] [PubMed] [Google Scholar]
- McElroy AE. Polycyclic aromatic hydrocarbon metabolism in the polychaete Nereis virens. Aquat. Toxicol. 1990;18:35–50. [Google Scholar]
- Menzel R, Bogaert T, Achazi R. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch. Biochem. Biophys. 2001;395:158–168. doi: 10.1006/abbi.2001.2568. [DOI] [PubMed] [Google Scholar]
- Menzel R, Rodel M, Kulas J, Steinberg CE. CYP35: xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005;438:93–102. doi: 10.1016/j.abb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Menzel R, Yeo HL, Rienau S, Li S, Steinberg CE, Sturzenbaum SR. Cytochrome P450s and short-chain dehydrogenases mediate the toxicogenomic response of PCB52 in the nematode Caenorhabditis elegans. J. Mol. Biol. 2007;370:1–13. doi: 10.1016/j.jmb.2007.04.058. [DOI] [PubMed] [Google Scholar]
- Meyer JN. QPCR: a tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology. Ecotoxicology. 2010;19:804–811. doi: 10.1007/s10646-009-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Lord CA, Yang XY, Turner EA, Badireddy AR, Marinakos S, Chilkoti A, Wiesner MR, Auffan M. Intracellular uptake and associated toxicity of silver nanoparticles in Caenorhabditis elegans. Aquat. Toxicol. 2010;100:140–150. doi: 10.1016/j.aquatox.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Miller LM, Hartman PS. The effects of benzo[a]pyrene (cough cough!) on C. elegans. Worm Breed. Gaz. 1998;15:43. [Google Scholar]
- Nelson DR. The cytochrome P450 homepage. Hum. Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil N, Rose A. DNA repair. In: Community TCER, editor. 2005. Available at: http://www.wormbook.org. Accessed October 20, 2010. [Google Scholar]
- Peterson RT, Nass R, Boyd WA, Freedman JH, Dong K, Narahashi T. Use of non-mammalian alternative models for neurotoxicological study. Neurotoxicology. 2008;29:546–555. doi: 10.1016/j.neuro.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Qin H, Zhai Z, Powell-Coffman JA. The Caenorhabditis elegans AHR-1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev. Biol. 2006;298:606–615. doi: 10.1016/j.ydbio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Rajini PS, Melstrom P, Williams PL. A comparative study on the relationship between various toxicological endpoints in Caenorhabditis elegans exposed to organophosphorus insecticides. J. Toxicol. Environ. Health A. 2008;71:1043–1050. doi: 10.1080/15287390801989002. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Le Bot N, Ahringer J, Aroian RV. Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev. Biol. 2003;3:8. doi: 10.1186/1471-213X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert K, Menzel R. Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere. 2005;61:229–237. doi: 10.1016/j.chemosphere.2005.01.077. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Styrishave B, Lobner-Olesen A, Andersen O. Marine invertebrate cytochrome P450: emerging insights from vertebrate and insect analogies. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006;143:363–381. doi: 10.1016/j.cbpc.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Saarikoski S, Rivera S, Hankinson O, Husgafvel-Pursiainen K. CYP2S1: a short review. Toxicol. Appl. Pharmacol. 2005;207:62–69. doi: 10.1016/j.taap.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Sancar A, Reardon JT. Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- Schäfer P, Müller M, Krüger A, Steinberg CEW, Menzel R. Cytochrome P450-dependent metabolism of PCB52 in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2009;488:60–68. doi: 10.1016/j.abb.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Schultz TW. Nonpolar narcosis: a review of the mechanism of action for baseline aquatic toxicity. In: Cowgill UM, editor; Cowgill UM, Williams LR, editors. Aquatic Toxicity and Hazard Assessment: Vol. 12, ASTM STP 1027. Philadelphia, PA: American Society for Testing and Materials; 1989. pp. 104–109. [Google Scholar]
- Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MV, Boyd WA, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. A discrete time model for the analysis of medium-throughput C. elegans growth data. PLoS ONE. 2009;4:e7018. doi: 10.1371/journal.pone.0007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprando RL, Olejnik N, Cinar HN, Ferguson M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a complex object parametric analyzer and sorter, and axenic liquid media. Food Chem. Toxicol. 2009;47:722–728. doi: 10.1016/j.fct.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stergiou L, Hengartner MO. Death and more: DNA damage response pathways in the nematode C elegans. Cell Death Differ. 2004;11:21–28. doi: 10.1038/sj.cdd.4401340. [DOI] [PubMed] [Google Scholar]
- Stewart HI, Rosenbluth RE, Baillie DL. Most ultraviolet-irradiation induced mutations in the nematode Caenorhabditis elegans are chromosomal rearrangements. Mutat. Res. 1991;249:37–54. doi: 10.1016/0027-5107(91)90131-7. [DOI] [PubMed] [Google Scholar]
- Sulston J. Cell lineage. In: Wood WB, editor. The Nematode Caenorhabitis elegans. pp. 123–155. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Valmas N, Ebert PR. Comparative toxicity of fumigants and a phosphine synergist using a novel containment chamber for the safe generation of concentrated phosphine gas. PLoS ONE. 2006;1:e130. doi: 10.1371/journal.pone.0000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ. Trends in hydrophobic organic contaminants in urban and reference lake sediments across the United States, 1970-2001. Environ. Sci. Technol. 2005;39:5567–5574. doi: 10.1021/es0503175. [DOI] [PubMed] [Google Scholar]
- Vineis P, Xun W. The emerging epidemic of environmental cancers in developing countries. Ann. Oncol. 2009;20:205–212. doi: 10.1093/annonc/mdn596. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Kobayashi K, Kato Y, Oda S, Abe R, Tatarazako N, Iguchi T. Transcriptome profiling in crustaceans as a tool for ecotoxicogenomics: Daphnia magna DNA microarray. Cell Biol. Toxicol. 2008;24:641–647. doi: 10.1007/s10565-008-9108-4. [DOI] [PubMed] [Google Scholar]
- Williams PL, Dusenbery DB. Aquatic toxicity testing using the nematode, Caenorhabditis elegans. Environ. Toxicol. Chem. 1990;9:1285–1290. [Google Scholar]