Abstract
Sulfotransferase isoform 1A1 (SULT1A1) is the most highly expressed hepatic sulfotransferase and is involved in the biotransformation of a wide variety of endo- and xenobiotics. A common single nucleotide polymorphism (SNP) in the coding region of SULT1A1, several proximal promoter SNPs, and copy number variation (CNV) are associated with altered enzymatic activity, but these variants do not fully account for the observed variation of SULT1A1 activity in human populations. In order to identify additional SNPs modulating SULT1A1 activity, we examined the 3′-untranslated region (UTR) of SULT1A1 in 97 liver samples. Direct sequencing revealed that two SNPs in the 3′-UTR (902A > G [rs6839] and 973C > T [rs1042157]) and one SNP in the 3′-flanking region (1307G > A [rs4788068]) were common. These SNPs are in absolute linkage disequilibrium with each other and in tight linkage with SULT1A1*1/2 (linkage coefficient D′ 0.83) and are significantly associated with SULT1A1 messenger RNA (p = 0.001, 0.029, 0.021) and enzymatic activity (p = 0.022, 0.012, 0.027). We then examined the collective effects of 3′-UTR SNPs, SULT1A1*1/2, and CNV on SULT1A1 activity in 498 Caucasian and 127 African-American subjects by haplotype analysis. This analysis revealed that SULT1A1*1/2 does not contribute to the variation in SULT1A1 enzymatic activity when the 3′-UTR SNPs are included in the statistical model. Two major haplotypes (ACG and GTA) were significantly correlated with SULT1A1 activity, and when stratified by copy number, the SULT1A1 3′-UTR SNPs remain significantly associated with SULT1A1 enzymatic activity in Caucasians, but not in African-Americans. Subsequent functional characterization revealed that a microRNA, miR-631, regulates SULT1A1 expression in a genotype-specific manner.
Keywords: SULT1A1, genotype, phenotype, pharmacogenetics
Sulfotransferase isoform 1A1 (SULT1A1) is a member of the sulfotransferase (SULT) family of phase II detoxification enzymes that catalyze the transfer of the sulfonyl group from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to nucleophilic groups of a variety of xenobiotic and endogenous compounds, thus increasing their solubility and facilitating their excretion (Jakoby and Ziegler, 1990). Several therapeutic agents, including acetaminophen, minoxidil, diethylstilbestrol, and 4-hydroxytamoxifen, are substrates for SULT1A1, and variability in the activity levels of the enzyme can markedly influence the efficacy of these drugs (Hildebrandt et al., 2009). For example, studies have demonstrated that the SULT1A1*1 high-activity allele is associated with much better overall survival in breast cancer patients receiving tamoxifen (Nowell et al., 2002, 2005). SULT1A1 activity is also involved in the activation of procarcinogens found in high-temperature cooked meats (reviewed in Glatt, 1997). Given the central role that SULT1A1 plays in drug metabolism, elucidation of the genetic determinants of SULT1A1 activity is essential for pharmacogenomic studies of therapeutic agents that are SULT1A1 substrates.
SULT1A1 is the most highly expressed SULT in the liver. SULT1A1 platelet activity has been demonstrated to correlate highly with activities in liver, intestinal tract, and brain (Campbell et al., 1987; Sebat et al., 2004; Sundaram et al., 1989), leading to the use of platelet enzyme as a surrogate for hepatic enzyme activity in human population studies. SULT1A1 activity varies several fold among individuals and shows a strong familial segregation, indicating that genetic factors play an important role in determining the enzymatic activity of SULT1A1 (Van Loon and Weinshilboum, 1984). A common single nucleotide polymorphism (SNP) in the coding region of SULT1A1 (638G > A, rs9282861, Arg213His, and SULT1A1*1/2) has been identified and is associated with decreased platelet enzymatic activity and thermostability (Nowell et al., 2000; Ozawa et al., 1998; Raftogianis et al., 1997). Another variant allele, SULT1A1*3 (667A > G, rs1801030, and Met223Val), is common in African-Americans but rare in Caucasians (Carlini et al., 2001). Four common and one rare SNP in the proximal promoter region, which are associated with platelet enzymatic activity (Ning et al., 2005), have also been reported and are in varying degrees of linkage disequilibrium with SULT1A1*1/2. Nonetheless, whether these SNPs are examined individually or in haplotypes, they account for only a small proportion of the variation observed in platelet activity.
SNPs have been the predominant type of variation explored in the human genome, but copy number variation (CNV) has been reported recently to be another important contributor to human genetic variation (Iafrate et al., 2004; Sebat, et al., 2004). Hebbring et al. (2007) demonstrated the presence of SULT1A1 gene deletions and duplications, providing an additional contributor to variability in the metabolic activity of this enzyme; CNVs also display ethnic differences. As with SNPs, CNV did not fully account for population variability in enzymatic activity; thus, it is possible as yet unidentified genetic variants have collective effects with CNV on the modulation of SULT1A1 activity.
In this study, we screened the 3′-untranslated region (UTR) of SULT1A1 in DNA from 97 human liver specimens to identify common SNPs. We identified and characterized two SNPs in the 3′-UTR and one SNP in 3′-flanking region (902A > G [rs6839] and 973C > T [rs1042157], and 1307G > A [rs4788068]) that were closely associated with both SULT1A1 expression and enzymatic activity. Haplotype analysis of the collective effects of functional genetic variants in the 3′-UTR regions, SULT1A1*1/2, and CNV on SULT1A1 enzymatic activity were then performed in 498 Caucasian and 127 African-American subjects. When stratified by copy number, SULT1A1 3′-UTR SNPs and the haplotypes ACG and GTA are significantly associated with SULT1A1 enzymatic activity in Caucasians, and the collective effect of CNV and 3′-UTR SNPs account for 21% of the variation in SULT1A1 activity in this population.
We then investigated the mechanism by which these SNPs could impact SULT1A1 expression. In silico analyses predicted that 973C > T influences the binding of miR-631 to the SULT1A1 3′-UTR. In vitro luciferase reporter assays and overexpression of microRNA (miRNA) inhibitors in ZR75-1, MCF7, and MCF10A breast cell lines confirm that SULT1A1 is a direct target of miR-631, and its messenger RNA (mRNA) is differentially regulated in an allele-specific manner.
MATERIALS AND METHODS
SNP identification and genotyping.
Human SULT1A1 genomic sequence (GenBank accession no. U52852) was used to design primers to amplify a 609-bp fragment of the 3′-UTR. The primer sequence was 5′-ggactggaagaccaccttc-3′ and 5′-tcaggcttgagtggattagc-3′. PCR was performed using 5 μl JumpStart REDTaq ReadyMix Reaction Mix (Sigma, St Louis, MO) in a total volume of 10 μl containing 3 ng genomic DNA and 0.5μM of each primer. PCR products were amplified using the following thermal cycling conditions: initial denaturation at 95°C for 4 min, 35 cycles of 94°C for 50 s, 60°C for 50 s, and 72°C for 1 min, followed by a final extension step of 10 min at 72°C. Genotype was determined by direct sequencing using the CEQ DTCS-Quick Start Sequencing Kit (Beckman Coulter, Inc. Brea, CA) and the CEQ8800 Genetic Analysis System. Genotyping for SULT1A1*1/2 was performed as previously described (Nowell et al., 2000).
Study subjects.
For our initial sequencing studies, 97 normal snap-frozen liver specimens were obtained from the U.S. Cooperative Human Tissue Network under an Institutional Review Board (IRB)–approved protocol. There were 43 female and 54 male subjects (age range 26–102 years, mean 59.0 years). Because only three subjects were African-Americans, we restricted our analyses to Caucasians.
For subsequent haplotype-phenotype studies, DNA and SULT1A1 platelet activity obtained from 625 healthy individuals who served as controls in breast and prostate cancer case-control studies in Arkansas was employed (Nowell et al., 2000, 2004). The population consisted of 254 females and 371 males (age range 32–89 years, mean 62.8 years). Of these subjects, 498 Caucasians and 127 African-Americans had both genotyping and phenotyping data available. The IRB at the University of Arkansas for Medical Sciences approved these study protocols.
Preparation of liver and platelet cytosols and SULT activity assay.
Liver cytosols were prepared from the frozen tissues. Briefly, liver tissue was minced with scissors and homogenized in homogenization buffer (0.25M sucrose, 50mM Tris-HCl, pH 7.8, 5mM EDTA, 20μM butylated hydroxytoluene, and 0.1mM dithiothreitol). The homogenates were centrifuged at 10,000 × g at 4°C for 10 min, and the resulting supernatant was subjected to ultracentrifugation at 100,000 × g for 1 h. The resulting cytosolic fraction was aliquoted, flash-frozen in liquid nitrogen, and stored at −70°C. Platelet cytosol was prepared as previously described (Frame et al., 2000). SULT1A1 enzymatic activity was determined using a colorimetric assay first described by Mulder et al., (1977) that was modified to a microtiter plate format (Frame et al., 2000). Activity was reported as nanomoles per minute per milligram protein.
Real-time PCR.
Liver DNA and RNA were extracted using AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA). Cellular RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, California) and reverse transcribed using SuperScript III First-Strand Synthesis System for reverse transcription (RT)-PCR Kit (Invitrogen) according to the manufacturer's instructions. SULT1A1 probe- and gene-specific primers were designed and purchased from Applied Biosystems (Foster City, CA). The primer sequences were as follows: forward 5′TGCCCGCAACGCAAA3′ and reverse 5′GGCCATGTGGTAGAAGTGGTAGT3′. SULT1A1 probe was FAM-5′ATGTGGCAGTTTCC3′. Custom β-actin and 18S probes were used as internal controls. Relative gene expression quantification for SULT1A1 was carried out in triplicate in an ABI 7900HT real-time PCR system (Applied Biosystems Inc.). Amplification of complementary DNA (cDNA) was performed using TaqMan Universal PCR Master Mix and initiated by the polymerase activation step for 10 min at 95°C. Amplification was obtained by 50 cycles of 15 s at 95°C with a 1-min annealing and extension step at 60°C.
For detection of miRNA expression in cells, miScript Reverse Transcription Kit (Qiagen) was used for cDNA synthesis. miScript SYBR Green PCR Kit (Qiagen), in combination with an miR-631–specific primer was used for mature miRNA detection. RNU6B was used as an internal control. Relative gene expression was determined using a previously described method (Livak and Schmittgen, 2001).
Western blots.
Cell lysates were separated by 12% SDS-polyacrylamide gel electrophoresis, and proteins were transferred onto a polyvinylidene fluoride membrane and probed with rabbit polyclonal anti-SULT1A1 antibody (Open Biosystems, Huntsville, AL) at 4°C overnight, and then the membrane was incubated with goat-anti-rabbit secondary antibody for 1 h before chemiluminescence detection using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL). β-Actin was also detected as a loading control using mouse monoclonal antibody (Sigma).
CNV assays.
SULT1A1 copy number determination was performed by real-time PCR in an ABI PRISM Sequence Detection System 7900 Instrument (Applied Biosystems) using the TaqMan Gene Expression Absolute Quantification Assay. A pair of unlabeled PCR primers, 5′-TGCCCGCAACGCAAA-3′ and 5′-GGCCATGTGGTAGAAGTGGTAGT-3′, and a FAM dye–labeled TaqMan MGB probe, 5′-ATGTGGCAGTTTCC-3′, were designed to specifically amplify SULT1A1. VIC dye–labeled TaqMan RNase P, which has two copies per haploid human genome, was used as a control. Amplification was 10 min of initial setup at 95°C, followed by 40 amplification cycles (15 s of denaturation at 95°C and 60 s of annealing/extension at 60°C). Each sample was examined in quadruplicate, and copy number was determined using Copycaller software (Applied Biosystems).
Cell culture and RNA degradation analysis.
The breast cancer cell lines ZR75-1, MCF7, and the mammary epithelial cell line MCF10A were obtained from the American Type Culture Collection (Rockville, MD) and were maintained at 37°C under 5% CO2. ZR75-1 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen). MCF7 cells were maintained in improved minimum essential media supplemented with 10% fetal bovine serum and 0.01 mg/ml bovine insulin (Invitrogen). MCF10A cells were cultured in complete mammary epithelium growth medium (Clonetics Corporate, Warrenale, PA).
For RNA degradation analysis, cells were seeded into six-well plates and grown to subconfluence, and then actinomycin D (Sigma) was added to a final concentration of 5 μg/ml to arrest de novo RNA synthesis. Cells were harvested at 3, 6, 9, and 12 h after actinomycin D treatment, and SULT1A1 mRNA was quantified by real-time PCR as described above. β-Actin mRNA level was monitored as internal control. All experiments were performed in triplicate.
Plasmid construction and luciferase assay.
To produce luciferase constructs containing 973C > T, the SULT1A1 3′-UTR was amplified using a pair of primers whose sequence was 5′-gcTCTAGAgaggggctcctggagtc-3′ and 5′-gcTCTAGAaaaaattggttttattttattttatttt-3′. PCR was performed with genomic DNA from subjects homozygous for the 973 CC or TT genotype to generate 973 C or 973 T luciferase constructs. The PCR product was digested using XbaI and ligated into pGL3-control vector (Promega Corporation, Madison, WI). The constructs were sequenced to assure proper orientation and authenticity in the vector.
Cells were plated in a 96-well plate and grown to 80–90% confluence. The firefly luciferase constructs (100 ng) were cotransfected with 30nM miR-631 mimics into MCF7 cells using Lipofectamine 2000 reagent. To monitor transfection efficiency, cells were cotransfected with 20 ng of the pRL-SV40 plasmid that encodes Renilla luciferase. Luminescence was measured 24 h after transfection using a dual-luciferase reporter assay system (Promega Corporation). All transfections were performed in triplicate, and data were analyzed by normalizing firefly luciferase activity to Renilla luciferase activity for each sample. Each construct was tested in three independent transfections.
Transfection of miR-631 inhibitor.
MiR-631 inhibitor was purchased from Ambion and transfected into cells with Lipofectamine 2000 (Invitrogen). The final concentration of inhibitor was 40nM. After 48 h, cells were harvested and miR-631 and SULT1A1 expression were measured. A nonspecific miRNA inhibitor was used as a negative control.
Statistical analysis.
Prior to data analysis, genotyping data were examined for possible genotyping errors by testing deviation from Hardy-Weinberg equilibrium. Linkage disequilibrium coefficients (D′) for each pair of SNPs were generated using the 2LD program (Zhao, 2004). ANOVA was carried out to identify SNPs that significantly influenced SULT1A1 expression and activity, with expression and activity as dependent variables. SULT1A1 expression was normalized with respect to 18S as an internal control. Functional SNPs (those significantly associated with expression or enzymatic activity) identified by preliminary screening were used to construct haplotypes. Frequencies and best pair of haplotypes for each individual were predicted using the software program PHASE (Stephens and Donnelly, 2003; Stephens et al., 2001). SULT1A1 expression and activity differences by race and gender were examined using Student's t-test. To evaluate the independent main effects of each of the functional SNPs and haplotypes of interest, multivariable ANOVA and regression analyses were carried out and partial sum of square, partial R2, regression coefficient (β), and p values were reported. Multivariable models were adjusted for age, race, and gender and the potential interactions between predictors of phenotype examined. None of the interaction terms constructed was found to be statistically significant and were thus removed from the ANOVA and regression models. Statistical significance was set at p < 0.05 (two sided) for individual test and Bonferroni-adjustment applied in significance level for multiple comparisons. Data preparation and analyses were performed using SAS software (version 9.2; SAS Institute, Inc., Cary, NC).
Similar statistical analysis procedures as those described above were adopted for DNA samples and platelet enzymatic activity from healthy individuals who served as controls in breast and prostate cancer case-control studies in Arkansas (n = 625). We examined whether the 3'-UTR SNPs, 902A INS> > G, 973C > T, and 1307G > A were independent from SULT1A1*1/2 in predicting SULT1A1 activity. We constructed haplotypes from the three functional SNPs in the 3′-UTR and introduced it as a class variable (four groups) together with SULT1A1*1/2 (three groups), copy number (four groups), and confounders (age, race, and gender) in the ANOVA model. The effects of each term included in the model were evaluated from partial sum of square and p value.
RESULTS
3′-UTR Allele Frequency and Linkage with SULT1A1*1/2
We screened SNPs in the 3′-UTR of SULT1A1 in 97 liver samples and characterized two SNPs (902A > G and 973C > T) in the 3′-UTR and one SNP (1307G > A) in the 3′-flanking region. The numbering of bases was designated relative to translation start site, excluding introns. When these SNPs were examined in healthy study subjects (n = 625), ethnic variation in allele frequency was evident. The variant allele frequencies are presented in Table 1. Genotype distributions of these SNPs conformed to Hardy-Weinberg equilibrium (p > 0.05).
TABLE 1.
Results of ANOVA and Linear Regression of Liver SULT1A1 Activity (Nanomoles per Minute per Milligram Protein) and mRNA with SULT1A1 3′-UTR and Coding Region Genotypes and Haplotypes
| Frequency | Partial sum of square | Partial R2 | β Coefficient | p Value | |
| SULT1A1 activitya | |||||
| Genotypes | |||||
| 902A > G | 0.42 | 3.03 | 0.06 | −0.27 | 0.022 |
| 973C > T | 0.45 | 3.63 | 0.07 | −0.30 | 0.012 |
| 1307G > A | 0.46 | 2.83 | 0.05 | −0.26 | 0.027 |
| 638 G > A | 0.42 | 3.24 | 0.06 | −0.27 | 0.017 |
| Haplotypesc | |||||
| ACGG | 0.54 | 2.9 | 0.05 | 0.26 | 0.025 |
| GTAA | 0.41 | 3.82 | 0.07 | −0.31 | 0.010 |
| Total model SS | 54.0 | ||||
| SULT1A1 mRNAb | |||||
| Genotypes | |||||
| 902A > G | 2.54 | 0.11 | −0.26 | 0.001 | |
| 973C > T | 1.27 | 0.05 | −0.19 | 0.029 | |
| 1307G > A | 1.43 | 0.06 | −0.20 | 0.021 | |
| 638 G > A | 1.91 | 0.08 | −0.21 | 0.007 | |
| Haplotypesc | |||||
| ACGG | 0.54 | 1.36 | 0.06 | 0.19 | 0.024 |
| GTAA | 0.41 | 1.70 | 0.07 | −0.21 | 0.011 |
| Total model SS | 24.2 | ||||
Note.Bold values indicate haplotypes are predominant and statistically significant. SS, sum of square.
Models with natural log of SULT1A1 activity as dependent variable were adjusted for age and gender.
Models with SULT1A1 mRNA as dependent variable were adjusted for age and gender.
Haplotypes were constructed from functional SNPs 902A > G, 973C > T, 1307G > A, or 638 G > A.
The linkage disequilibrium analysis as quantified by the linkage coefficient D′ showed that SNPs in 3′-UTR and 3′-flanking region are in absolute linkage disequilibrium, and they are also in tight linkage with SULT1A1*1/2 (linkage coefficient D′ = 0.83–0.87, data not shown).
Association of SULT1A1 SNPs in the 3′-UTR Regions and SULT1A1*1/2 SNP with SULT1A1 Enzymatic Activity in Liver
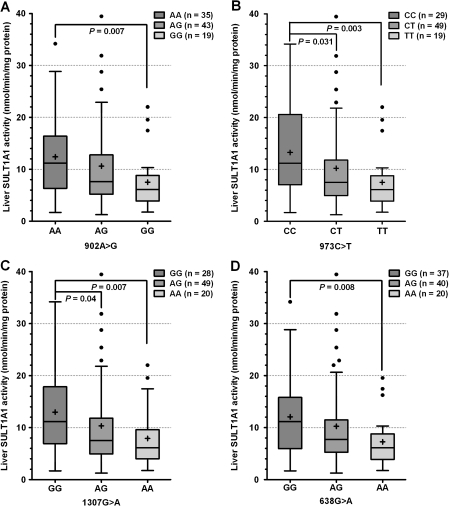
To investigate whether the SULT1A1 3′-UTR SNPs influence SULT1A1 enzymatic activity in human liver cytosols, we performed ANOVA analysis to examine the association of each individual SNP with SULT1A1 activity. After adjusting for age and gender, 902A > G, 973C > T, 1307G > A, and 638G > A (i.e., SULT1A1*1/2) were significantly associated with SULT1A1 activity (p = 0.022, 0.012, 0.027, and 0.017, respectively; Table 1). As shown in Figure 1, 3′-UTR variant homozygous genotypes 902GG, 973TT, 1307AA, and 638AA are associated with decreased liver SULT1A1 activity. Pairwise comparison revealed significant differences in SULT1A1 activity between the homozygous genotype groups (p < 0.05). SULT1A1 activity also differed by gender (13.1 ± 9.0, female, 9.1 ± 7.1, male, p = 0.022). After adjusting for age and gender, 902A > G, 973C > T, 1307G > A, and 638G > A individually accounted for 6, 7, 5, and 6% of the variation in SULT1A1 enzymatic activity in liver.
FIG. 1.
Box plot analysis of liver SULT1A1 activity by SULT1A1 SNPs. The plus shape sign and line inside each box indicate mean and median, and the upper and lower limits of the box are 75th and 25th percentiles, respectively, and the vertical bars above and below show the maximum and minimum values, respectively. The solid circles outside the box are outlier values. p Values are shown only for pairs of genotype whose mean level of activity was statistically significant.
Association of SULT1A1 3′-UTR SNPs and SULT1A1*1/2 with SULT1A1 Gene Expression in Human Liver
Because SULT1A1 SNPs in the 3′-UTR have a significant effect on SULT1A1 enzymatic activity, we hypothesized that decreased SULT1A1 activity is because of reduced SULT1A1 expression in subjects harboring variant alleles. To test this hypothesis, we extracted RNA from liver samples and performed real-time PCR to detect SULT1A1 mRNA level normalized by 18S. ANOVA indicated that 902A > G, 973C > T, 1307G > A, and 638G > A were closely associated with SULT1A1 mRNA expression (p = 0.001, 0.029, 0.021, and 0.007, respectively; Table 1). Similarly, subjects with rare homozygous genotypes 902GG, 973TT, 1307AA, and 638AA had reduced SULT1A1 mRNA (p < 0.05, data not shown). Spearman correlation analysis indicated that liver SULT1A1 activity is closely associated with its expression (r = 0.524, p < 0.0001). After adjusting for age and gender, 902A > G, 973C > T, 1307G > A, and 638G > A individually accounted for 11, 5, 6, and 8% of the variation in SULT1A1 mRNA expression in liver.
SULT1A1 3′-UTR SNPs Vary by Ethnicity and Are Better Predictors of Platelet SULT1A1 Activity than SULT1A1*1/2
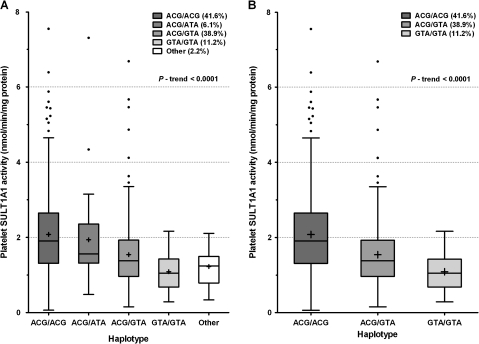
We then examined the distribution of the SNPs and their impact on SULT1A1 activity in a large population of healthy Caucasian and African-American individuals. The demographics of the study population are presented in the “Materials and Methods” section. To determine the significance of 3′-UTR SNPs on SULT1A1 activity, haplotype analysis of the collective effects of functional genetic variants in the 3′-UTR and SULT1A1*1/2 on SULT1A1 enzymatic activity was then performed in 498 Caucasian and 127 African-American subjects. African-Americans exhibited a 1.5-fold increase in basal platelet SULT1A1 activity (mean ± SD) compared with Caucasians (2.40 ± 1.40 vs. 1.60 ± 0.96). In agreement with the studies using human liver cytosols, SNPs 902A > G, 973C > T, 1307G > A, and 638G > A were closely associated with platelet SULT1A1 activity (Table 2). Haplotype construction demonstrated that ACGG and GTAA are the predominant haplotypes, with frequencies of 57 and 32%, respectively, and account for singly 8 and 7%, respectively, of the observed variation in SULT1A1 activity in Caucasians (Table 2). Most importantly, when the 3′-UTR haplotypes were taken into account, the SULT1A1*1/2 SNP had no significant influence on SULT1A1 enzymatic activity (p = 0.60). After adjustment for age, gender, race, and copy number, only the 3′-UTR haplotypes had a significant influence on SULT1A1 activity (p = 0.0001, Table 3). In fact, when we examined the 3′-UTR haplotypes in subjects whose phenotype and SULT1A1*1/*2 genotype were discordant (i.e., low activity when homozygous for the high-activity allele), phenotype was concordant with the 3′-UTR haplotype. Thus, the 638G > A SNP was dropped from the haplotype construction, and the haplotypes of the 3′-UTR and flanking region remain closely associated with SULT1A1 activity variation (p trend < 0.0001, Fig. 2). The predominant haplotypes ACG/ACG, ACG/GTA, and GTA/GTA are significantly associated with a trend of diminishing SULT1A1 activity (Fig. 2B). It is interesting to note that in African-Americans, the 3′-UTR SNPs, but not SULT1A1*1/2, were associated with platelet SULT1A1 activity (Table 2), which further highlights the important role of 3′-UTR SNPs in the determination of SULT1A1 activity.
TABLE 2.
Results of ANOVA and Linear Regression of Platelet SULT1A1 Activity (Nanomoles per Minute per Milligram Protein) with SULT1A1 3′-UTR and Coding Region Genotypes and Haplotypes Stratified by Race
| Frequency | Partial sum of square | Partial R2 | β Coefficient | p Value | |
| Caucasians | |||||
| Genotypes | |||||
| 902A > G | 0.35a | 16.7 | 0.09 | −0.27 | < 0.0001 |
| 973C > T | 0.39a | 15.1 | 0.08 | −0.25 | < 0.0001 |
| 1307G > A | 0.39a | 15.4 | 0.08 | −0.26 | < 0.0001 |
| 638 G > A | 0.36a | 13.2 | 0.07 | −0.23 | < 0.0001 |
| Haplotypesb | |||||
| ACGG | 0.57 | 14.2 | 0.08 | 0.24 | < 0.0001 |
| GTAA | 0.32 | 13.5 | 0.07 | −0.25 | < 0.0001 |
| GTAG | 0.026 | 2.3 | 0.01 | −0.27 | 0.012 |
| Total model SS | 181.5 | ||||
| African-Americans | |||||
| Genotypes | |||||
| 902A > G | 0.20a | 2.2 | 0.04 | −0.21 | 0.035 |
| 973C > T | 0.22a | 2.4 | 0.05 | −0.22 | 0.023 |
| 1307G > A | 0.22a | 2.4 | 0.05 | −0.22 | 0.023 |
| 638 G > A | 0.24a | 1.2 | 0.02 | −0.12 | 0.25 |
| Haplotypesb | |||||
| ACG | 0.78 | 2.3 | 0.04 | 0.22 | 0.027 |
| GTA | 0.20 | 2.1 | 0.04 | −0.21 | 0.042 |
| Total model SS | 52.3 | ||||
Note. Bold values indicate haplotypes are predominant and statistically significant. Models with natural log of SULT1A1 activity as dependent variable were adjusted for age and gender.
Minor allele frequency.
Haplotypes were constructed from functional SNPs 902A > G, 973C > T, 1307G > A, or 638 G > A.
TABLE 3.
Results of ANOVA of Platelet SULT1A1 Activity (Nanomoles per Minute per Milligram Protein) with SULT1A1 Coding Region Genotype, 3′-UTR Haplotype, and SULT1A1 Copy Number, All Subjects
| Partial sum of square | Partial R2 | F value | Pr > F | |
| 638G > A | 0.33 | < 0.01 | 0.51 | 0.60 |
| Haplotypea | 6.7 | 0.03 | 6.94 | 0.0001 |
| Copy numbera | 28.5 | 0.12 | 29.50 | < 0.0001 |
| Total model SS | 231.5 |
Note. Models with natural log of SULT1a1 activity as dependent variable were adjusted for age, gender, and race. Pr, probability.
Haplotype and copy number variables were in four categories, i.e., haplotype: ACG/ACG (41.6%), ACG/GTA (38.9%), GTA/GTA (11.2%), and other (ACG/ACG, ACG/ATA, ATA/ATA, ATA/GTA, or GCG/GTA) (8.3%); copy numbers one (12.6%), two (60.2%), three (22.9%), and four or more (4.3%).
FIG. 2.
Distributional analysis of platelet SULT1A1 activity by haplotypes (3'-UTR SNPs) in all subjects (n = 625). Contribution of (A) all haplotypes and (B) predominant haplotypes to platelet SULT1A1 activity.
Collective Effect of SULT1A1 3′-UTR SNPs and CNV on SULT1A1 Activity
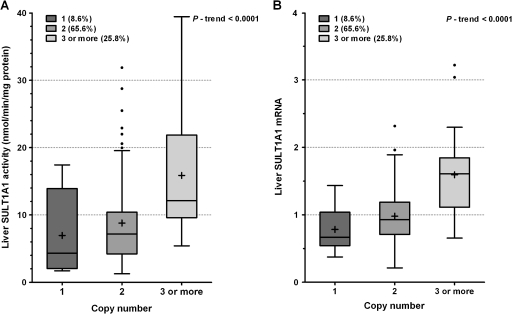
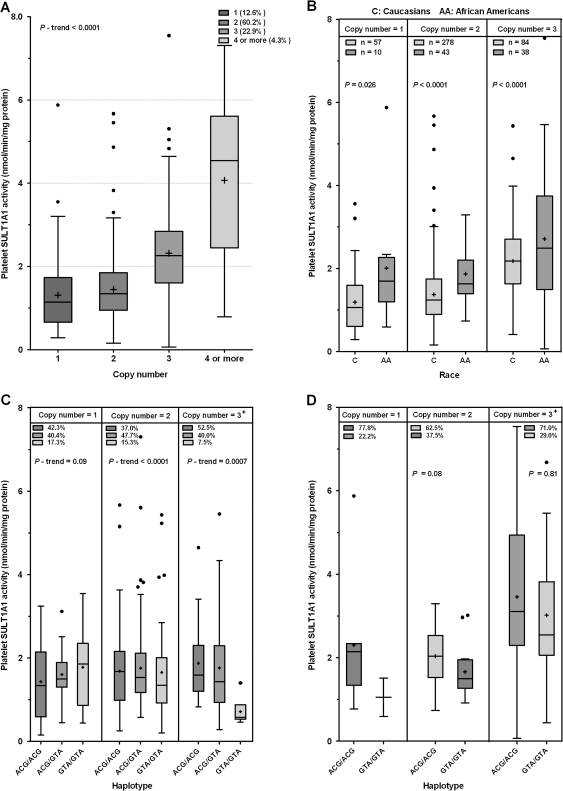
CNVs have recently been reported to have a significant influence on SULT1A1 activity (Hebbring et al., 2007). We determined CNV in the 97 liver samples, and the frequencies of SULT1A1 CNVs (one, two, and three or more) were 8.6, 65.6, and 25.8%, respectively, in Caucasians. SULT1A1 CNV is significantly associated with SULT1A1 activity and expression (p trend < 0.0001, Fig. 3). Pairwise comparison indicated that SULT1A1 activity is higher in subjects carrying higher copy number (three or more) than those carrying lower copy numbers (one or two) (p < 0.05). The collective effects of SULT1A1 3′-UTR SNPs and CNV on SULT1A1 activity were performed in 498 Caucasian and 127 African-American subjects. As expected, copy number was closely associated with SULT1A1 activity (p trend < 0.0001, Fig. 4A), and there were ethnic differences in copy number frequency distribution between Caucasians and African-Americans. Copy number one, two, three, and four or more frequencies were 13.2, 64.5, 19.5, and 2.8%, respectively, in Caucasians, whereas frequencies in African-American subjects were 9.8, 42.2, 37.2, and 10.8%, respectively, which can partially account for the higher platelet SULT1A1 activity observed in African-Americans compared with Caucasians. However, when stratified by copy number (Fig. 4B), significant differences in SULT1A1 activity between Caucasians and African-Americans among copy number one, two, and three groups (p = 0.026, p < 0.0001, p < 0.0001, respectively) persist, indicating that copy number alone cannot fully account for the ethnic differences in SULT1A1 activity. When stratified by copy number, haplotypes ACG/ACG, ACG/GTA, and GTA/GTA demonstrated a trend of decreasing SULT1A1 activity (p trend is shown in Figs. 4C and 4D). In Caucasians, these haplotypes were significantly associated with SULT1A1 activity among CNV two and three groups. In African-American subjects, these haplotypes did not appear to be associated with SULT1A1 activity when stratified by SULT1A1 CNV (possibly because of small numbers); however, they display the same trend as the Caucasian study subjects. We further assessed the additive effect of haplotypes and SULT1A1 CNV on phenotypic variation. As shown in Table 4, SULT1A1 CNV alone can account for 13% of SULT1A1 platelet enzymatic activity variation; when the 3'-UTR haplotypes are added to the model, the additive effects of both account for 21% of SULT1A1 activity variation in Caucasians.
FIG. 3.
Box plot analysis of liver SULT1A1 activity and SULT1A1 mRNA by CNV. Both SULT1A1 activity (A) and mRNA expression (B) are associated with SULT1A1 CNV.
FIG. 4.
Box plot analysis of the association of SULT1A1 3′-UTR and SULT1A1*1/2 with platelet SULT1A1 activity when stratified by CNV in 498 Caucasian and 127 African-American subjects. (A) CNV is associated with SULT1A1 activity in 625 subjects. (B) Ethnic difference of SULT1A1 activity stratified by CNV. (C and D) Association of SULT1A1 predominant haplotypes with SULT1A1 activity stratified by SULT1A1 CNV in Caucasians (C) and African-Americans (D).
TABLE 4.
Results of ANOVA of Platelet SULT1A1 Activity (Nanomoles per Minute per Milligram Protein) with 3′-UTR Haplotype and Copy Number Stratified by Race
| Partial sum of square | Partial R2 | F value | Pr > F | |
| Caucasians | ||||
| Haplotypea | 13.2 | 0.08 | 14.48 | < 0.0001 |
| Copy numbera | 21.2 | 0.13 | 35.02 | < 0.0001 |
| Total model SS | 167.0 | |||
| African-Americans | ||||
| Haplotypeb | 2.3 | 0.05 | 1.69 | 0.18 |
| Copy numberb | 3.3 | 0.07 | 3.55 | 0.033 |
| Total model SS | 49.1 | |||
Note. Models with natural log of SULT1A1 activity as dependent variable were adjusted for age, gender, and race. Pr, probability.
Haplotype and copy number variables were in four and three categories, respectively. Haplotype: ACG/ACG (36.8%), ACG/GTA (40.8%), GTA/GTA (13.1%), and other (ACG/ACA, ACG/ATA, ATA/ATA, ATA/GTA, or GCG/GTA) (9.4%); copy numbers one (13.2%), two (64.5%), and three or more (22.3%).
Haplotype and copy number variables were in three categories. Haplotype: ACG/ACG (60.6%), GTA/GTA (31.5%), and other (ACG/GTA, ACG/ACA, ACG/ATA, ATA/ATA, ATA/GTA, or GCG/GTA) (7.9%); copy numbers one (9.8%), two (42.2%), and three or more (48.0%).
SULT1A1 Degradation Profile and miRNA Regulation
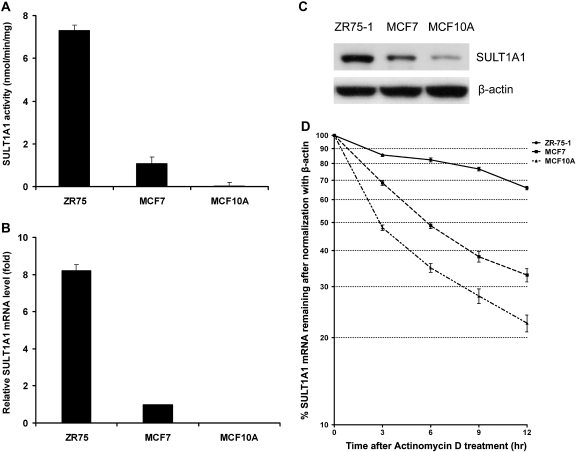
To elucidate the mechanism by which SULT1A1 3′-UTR variant alleles confer low SULT1A1 activity, we screened SULT1A1 3′-UTR genotypes in breast cell lines, all of which were derived from Caucasians. ZR75-1 possesses homozygous common alleles for all three SNPs; MCF7 has heterozygous alleles, whereas MCF10A is homozygous for the three variant alleles. The ZR75-1 cell line has four copies of SULT1A1, whereas MCF7 and MCF10A both have three copies. As expected, ZR75-1 cells exhibited the highest SULT1A1 expression and activity and MCF10A the lowest, which is consistent with our genotype-phenotype correlation in the population study (Figs. 5A and 5B). Therefore, we selected ZR75-1, MCF7, and MCF10A as cell models to explore the regulation of SULT1A1 in vitro. We hypothesized that cells with different genotypes in the 3′-UTR will have different patterns of SULT1A1 mRNA degradation, thereby resulting in different SULT1A1 expression levels. As expected, the relative amounts of expressed protein corresponded to levels of mRNA and enzymatic activity of the cell lines (Fig. 5C). To confirm this, we pretreated cells with actinomycin D to inhibit de novo RNA synthesis, and then cells were harvested, and mRNA levels were quantified at different time points (Fig. 5D). Results are presented as the percentages of SULT1A1 mRNA remaining after normalization with β-actin. SULT1A1 mRNA in ZR75-1 cells was fairly stable for 12 h, whereas SULT1A1 had a half-life of approximately 6 and 3 h in MCF7 and MCF10A cells, respectively. Thus, SULT1A1 expression differences may be because of different degradation profiles, with these profiles being genotype dependent.
FIG. 5.
Varying levels of SULT1A1 activity and expression in breast cells carrying different genotypes. (A) Colorimetric determination of SULT1A1 activity in breast cells ZR75-1, MCF7, and MCF10A. (B) Real-time PCR was used to detect endogenous SULT1A1 expression in ZR75-1, MCF7, and MCF10A cells. (C) Western blot was performed to examine SULT1A1 protein level in ZR75-1, MCF7, and MCF10A cells. (D) ZR75-1, MCF7, and MCF10A cells were treated with 5 μg/ml actinomycin D for the indicated time. SULT1A1 mRNA level was measured by RT-quantitative PCR, normalized to β-actin, and plotted on a logarithmic scale to show the remaining mRNA relative to its initial abundance. Values reported are mean ± SD of triplicate experiments.
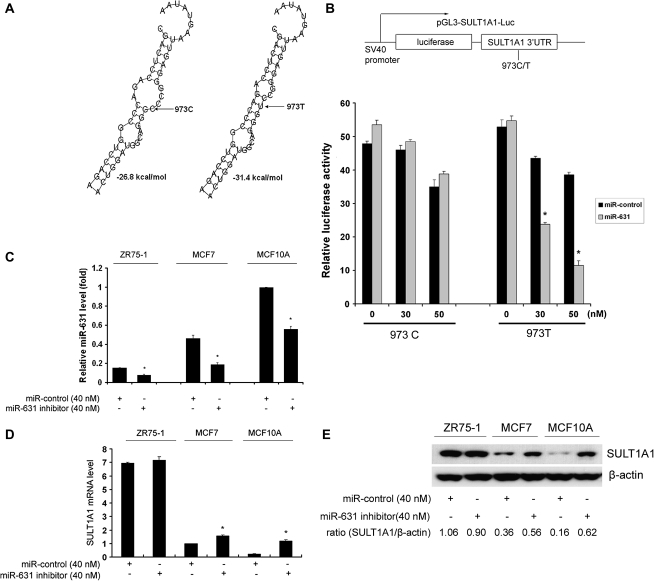
We next examined the potential interaction of the 3′-UTR SNPs with miRNA. MicroInspector (Rusinov et al., 2005) and RNAhybrid (Rehmsmeier et al., 2004) programs predicted that miR-631 would downregulate SULT1A1 expression more robustly in subjects carrying the 973T allele than in those with the C allele, with free energy decreased from −26.8 to −31.4 kcal/mol (Fig. 6A). SULT1A1 3′-UTR with 973C or T allele was cloned into pGL3 control vector and cotransfected with miR-631 into MCF7 cells. The T allele reduced luciferase activity relative to the C allele in a miR-631 dose-dependent manner, which suggests that 973C > T allele affects the binding of miR-631 to the SULT1A1 3′-UTR (Fig. 6B). Furthermore, miR-631 inhibitor was transiently transfected into ZR75-1, MCF7, and MCF10A cells. As shown in Figure 6C, this inhibitor was able to significantly decrease endogenous levels of miR-631 in each cell type. MiR-631 inhibitor enhanced both SULT1A1 mRNA and protein levels in MCF7 and MCF10A cells (Figs. 6D and 6E). However, this inhibitor had minimal effect on SULT1A1 expression in ZR75-1 cells, which would be expected as this cell line has no variant alleles.
FIG. 6.
MiR-631 differentially regulates SULT1A1 in allele-specific manner at 973C > T. (A) In silico analysis of the pairing of miR-631 to the binding site in the 3′-UTR of SULT1A1. The C to T allele change decreased free energy from −26.8 to −31.4 kcal/mol. (B) Luciferase activity in MCF7 cells transfected with pGL3-SULT1A1-973C or pGL3-SULT1A1-973T and cotransfected with variable doses of miR-631 mimics or miRNA negative control. All transfections were performed in triplicate, and relative luciferase activity was analyzed by normalizing firefly luciferase activity to Renilla luciferase activity for each sample. Asterisk denotes p < 0.05 compared with control. (C) ZR75-1, MCF7, and MCF10A cells were transfected with 40nM miR-631 inhibitor, and after 48 h, miR-631 RNA levels were determined. (D) mRNA levels for SULT1A1 were also determined, and (E) protein was extracted for Western blot to measure SULT1A1 protein level. β-Actin was used as a loading control. The density ratio of SULT1A1 versus β-actin was also indicated.
To further investigate the possible regulation of SULT1A1 mRNA by miR-631 and expression in a genotype-dependent manner, 97 liver samples were used to detect the expression level of miR-631, and its correlation with SULT1A1 was explored. MiR-631 was weakly negatively associated with SULT1A1 expression in liver (r = −0.19, p = 0.06); however, miR-631 expression was not influenced by SULT1A1 genotype. Thus, variant alleles in the SULT1A1 3′-UTR affect the binding ability of miR-631 to SULT1A1 but not the abundance of miR-631 (data not shown).
DISCUSSION
Genetic variation in SULT1A1 is of considerable interest, given the key role this enzyme plays in the metabolism of endo- and xenobiotics, procarcinogen activation/detoxification, and the disposition of therapeutic agents. Early family studies in twins (Reveley et al., 1982), as well as family studies (Price et al., 1988), showed that SULT1A1 activity was highly heritable and variable, raising the possibility of a major gene polymorphism. Other studies suggested that there were allelic forms of SULT1A1 whose activity could be because of thermal stability (Price et al., 1989), that the variability in activity was a result of differing levels of protein expressed (Jones et al., 1993), and the genetic polymorphism, when identified, would impact protein expression. In 1997, SULT1A1*1/*2 was identified and its association with enzymatic activity and thermostability was established (Raftogianis et al., 1997). However, the low-activity SULT1A1*2 allele was not consistently associated with hepatic SULT1A1 enzymatic activity (Raftogianis et al., 1999), and sulfating activity determinations using recombinant SULT1A1*1 and SULT1A1*2 failed to demonstrate an appreciable deficiency in [35S]PAPS-dependent sulfation of p-nitrophenol for the *2 enzyme (Ozawa et al., 1999). Genotype-phenotype studies showed that although there was a significant association between SULT1A1 genotype and platelet enzymatic activity, the percentage of variability attributable to the genotype was modest (Nowell et al., 2000). For this reason, we and others continued to search for other genetic modifiers of activity. Genetic variants in the proximal promoter region, which were in varying degrees of linkage with SULT1A1*1/*2, were reported (Ning et al., 2005), as well as CNVs (Hebbring et al., 2007). These variants are all associated with SULT1A1 enzymatic activity, with CNV accounting for the largest proportion of change, but again, the influence of the variants on phenotype is relatively modest.
In this study, we identified distal promoter SNPs that had a negligible effect on SULT1A1 activity compared with the SNPs in 3′-UTR. When these SNPs were included in modeling studies, they did not produce a change in the model and, thus, were not included in this study (data not shown). Haplotype activity analyses showed that haplotype GTAG (constructed from 902A > G, 973C > T, 1307G > A, and 638G > A) was associated with low platelet SULT1A1 activity (β coefficient = −0.25, p = 0.014), even though the final “G” in the haplotype corresponds to the high-activity SULT1A1*1 allele. Regardless of whether the haplotype was GTAG or GTAA, both were associated with low enzymatic activity. Furthermore, multivariate analysis indicated that when the 3′-UTR haplotypes were taken into account, the SULT1A1*2 allele had no significant influence on SULT1A1 enzymatic activity. Thus, the association of SULT1A1*1/*2 with SULT1A1 enzymatic activity is because of the tight linkage between this SNP and the 3′-UTR SNPs, which would account for the lack of effect of this SNP on 4-nitrophenol sulfation described in the earlier recombinant enzyme studies.
Variability in gene copy number is another source of genetic variation throughout the human genome (Conrad et al., 2006; Hinds et al., 2006) and has been observed in genes encoding drug-metabolizing enzymes (Johansson et al., 1993). Hebbring et al. (2007) demonstrated that SULT1A1 CNV displays ethnic differences in that 26% of Caucasians and 63% of the African-American subjects studied had more than two alleles. They further documented that the variability in SULT1A1 activity in platelet and liver samples was best explained by gene copy number differences. Our results are consistent with this finding when CNV is examined independently. However, addition of SULT1A1 3′-UTR SNPs to the analysis significantly enhances the model in that even when stratified by copy number, SULT1A1 3′-UTR SNPs remain significantly associated with enzymatic activity, whereas the SULT1A1*1/*2 allele does not. SULT1A1 CNV independently accounts for 13% of the variation in SULT1A1 platelet enzymatic activity, but the additive effects of SULT1A1 CNV and 3′-UTR haplotypes account for 21% of SULT1A1 activity variation, at least in Caucasians. As with other SNPs that have been described in SULT1A1, the impact of these SNPs in the African-American study subjects is less clear, although this may be because of the smaller number of African-Americans in this study. Even though the associations were not as strong as they were for Caucasians, the direction of the trend was the same. It is possible that there are other SNPs in unexplored intronic regions that have functional effects on SULT1A1 activity, but it is likely that a large proportion of the variability observed in human populations is because of environmental factors such as inhibition of activity by dietary constituents, many of which are known to be potent inhibitors of SULT1A1 activity, at least in in vitro studies (Ghazali and Waring, 1999; Pacifici, 2004; Vietri et al., 2003; Waring et al., 2008). Additionally, Marazziti and colleagues have reported variation in SULT1A1 activity in platelets that can be influenced by seasonal rhythms in a gender-specific manner (Marazziti et al., 1995, 1998). However the biological basis of this association is unclear.
MiRNAs are small noncoding RNAs, about 22 nucleotides long, that play important regulatory roles in animals and plants by targeting mRNA transcripts for cleavage or translational repression and regulate gene expression by binding to the 3′-UTR region of the target gene (Bartel and Chen, 2004). Functional polymorphisms in 3′-UTRs may interfere with the binding of miRNAs, leading to protein overproduction or decreased translation (Mishra et al., 2008). We hypothesized that the reduced expression of SULT1A1 in subjects harboring 902G and 973T alleles might be attributed to the high affinity of some miRNA to its binding site in 3′-UTR. To test this hypothesis, we selected three breast cell lines with typical SULT1A1 3′-UTR genotypes that exhibit different SULT1A1 expression and activity profiles. Although the genetic background of the cells is not exactly the same, all three cell lines are derived from Caucasian women, consistent with our SULT1A1 genotype-phenotype analysis, where the effect of genotype on activity is clear. The SULT1A1 degradation profile in these cells was consistent with the observed endogenous SULT1A1 expression and activity, suggesting that 3′-UTR genotype plays a key role in the modulation of SULT1A1 stability and degradation, therefore contributing to variation of SULT1A1 activity.
MicroInspector and RNAhybrid programs predicted that miR-631 would downregulate SULT1A1 at the 973T allele more strongly than the C allele, with free energy decreased from −26.8 to −31.4 kcal/mol. Luciferase reporter assays displayed the differential regulation of SULT1A1 by miR-631 at 973C > T allele, and transfection of miR-631 inhibitor into breast cells with different genotype characteristics of SULT1A1 3′-UTR confirmed that miR-631 directly regulates SULT1A1 expression in an allele-specific manner. In order to investigate whether miR-631 regulated SULT1A1 expression in tissue, real-time PCR was performed in 97 liver samples to detect miR-631 expression. MiR-631 was weakly associated with SULT1A1 expression in liver (r = −0.19, p = 0.06), suggesting that miR-631 plays a role in regulating SULT1A1 expression in human samples.
In summary, we identified SULT1A1 3′-UTR SNPs that predict SULT1A1 activity in liver and platelets. More importantly, we demonstrated for the first time that SULT1A1 is differentially regulated by miR-631 depending on the variant allele at 973C > T, providing new insight into the mechanism of SULT1A1 regulation.
FUNDING
National Cancer Institute at the National Institutes of Health (R01CA128897); National Center for Research Resources (award number 1UL1RR029884); Susan G. Komen for the Cure (BCTR0707584).
References
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Campbell NR, Van Loon JA, Weinshilboum RM. Human liver phenol sulfotransferase: assay conditions, biochemical properties and partial purification of isozymes of the thermostable form. Biochem. Pharmacol. 1987;36:1435–1446. doi: 10.1016/0006-2952(87)90108-0. [DOI] [PubMed] [Google Scholar]
- Carlini EJ, Raftogianis RB, Wood TC, Jin F, Zheng W, Rebbeck TR, Weinshilboum RM. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics. 2001;11:57–68. doi: 10.1097/00008571-200102000-00007. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Frame LT, Ozawa S, Nowell SA, Chou HC, DeLongchamp RR, Doerge DR, Lang NP, Kadlubar FF. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab. Dispos. 2000;28:1063–1068. [PubMed] [Google Scholar]
- Ghazali RA, Waring RH. The effects of flavonoids on human phenolsulphotransferases: potential in drug metabolism and chemoprevention. Life Sci. 1999;65:1625–1632. doi: 10.1016/s0024-3205(99)00423-3. [DOI] [PubMed] [Google Scholar]
- Glatt H. Sulfation and sulfotransferases 4: bioactivation of mutagens via sulfation. FASEB J. 1997;11:314–321. doi: 10.1096/fasebj.11.5.9141497. [DOI] [PubMed] [Google Scholar]
- Hebbring SJ, Adjei AA, Baer JL, Jenkins GD, Zhang J, Cunningham JM, Schaid DJ, Weinshilboum RM, Thibodeau SN. Human SULT1A1 gene: copy number differences and functional implications. Hum. Mol. Genet. 2007;16:463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- Hildebrandt M, Adjei A, Weinshilboum R, Johnson JA, Berlin DS, Klein TE, Altman RB. Very important pharmacogene summary: sulfotransferase 1A1. Pharmacogenet. Genomics. 2009;19:404–406. doi: 10.1097/FPC.0b013e32832e042e. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jakoby WB, Ziegler DM. The enzymes of detoxication. J. Biol. Chem. 1990;265:20715–20718. [PubMed] [Google Scholar]
- Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjoqvist F, Ingelman-Sundberg M. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Roberts RC, Coughtrie MW. The human phenolsulphotransferase polymorphism is determined by the level of expression of the enzyme protein. Biochem. J. 1993;296(Pt 2):287–290. doi: 10.1042/bj2960287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marazziti D, Palego L, Mazzanti C, Silvestri S, Cassano GB. Human platelet sulfotransferase shows seasonal rhythms. Chronobiol. Int. 1995;12:100–105. doi: 10.3109/07420529509064505. [DOI] [PubMed] [Google Scholar]
- Marazziti D, Palego L, Rossi A, Cassano GB. Gender-related seasonality of human platelet phenolsulfotransferase activity. Neuropsychobiology. 1998;38:1–5. doi: 10.1159/000026509. [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Mishra PJ, Banerjee D, Bertino JR. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–858. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- Mulder GJ, Hinson JA, Gillette JR. Generation of reactive metabolites of N-hydroxy-phenacetin by glucoronidation and sulfation. Biochem. Pharmacol. 1977;26:189–196. doi: 10.1016/0006-2952(77)90301-x. [DOI] [PubMed] [Google Scholar]
- Ning B, Nowell S, Sweeney C, Ambrosone CB, Williams S, Miao X, Liang G, Lin D, Stone A, Ratnasinghe DL, et al. Common genetic polymorphisms in the 5'-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet. Genomics. 2005;15:465–473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- Nowell S, Ambrosone CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF, Lang NP. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10:789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- Nowell S, Ratnasinghe DL, Ambrosone CB, Williams S, Teague-Ross T, Trimble L, Runnels G, Carrol A, Green B, Stone A, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol. Biomarkers Prev. 2004;13:270–276. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J. Natl. Cancer Inst. 2002;94:1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res. Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- Ozawa S, Shimizu M, Katoh T, Miyajima A, Ohno Y, Matsumoto Y, Fukuoka M, Tang YM, Lang NP, Kadlubar FF. Sulfating-activity and stability of cDNA-expressed allozymes of human phenol sulfotransferase, ST1A3*1 ((213)Arg) and ST1A3*2 ((213)His), both of which exist in Japanese as well as Caucasians. J. Biochem. 1999;126:271–277. doi: 10.1093/oxfordjournals.jbchem.a022445. [DOI] [PubMed] [Google Scholar]
- Ozawa S, Tang YM, Yamazoe Y, Kato R, Lang NP, Kadlubar FF. Genetic polymorphisms in human liver phenol sulfotransferases involved in the bioactivation of N-hydroxy derivatives of carcinogenic arylamines and heterocyclic amines. Chem. Biol. Interact. 1998;109:237–248. doi: 10.1016/s0009-2797(97)00135-x. [DOI] [PubMed] [Google Scholar]
- Pacifici GM. Inhibition of human liver and duodenum sulfotransferases by drugs and dietary chemicals: a review of the literature. Int. J. Clin. Pharmacol. Ther. 2004;42:488–495. doi: 10.5414/cpp42488. [DOI] [PubMed] [Google Scholar]
- Price RA, Cox NJ, Spielman RS, Van Loon JA, Maidak BL, Weinshilboum RM. Inheritance of human platelet thermolabile phenol sulfotransferase (TL PST) activity. Genet. Epidemiol. 1988;5:1–15. doi: 10.1002/gepi.1370050102. [DOI] [PubMed] [Google Scholar]
- Price RA, Spielman RS, Lucena AL, Van Loon JA, Maidak BL, Weinshilboum RM. Genetic polymorphism for human platelet thermostable phenol sulfotransferase (TS PST) activity. Genetics. 1989;122:905–914. doi: 10.1093/genetics/122.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem. Biophys. Res. Commun. 1997;239:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem. Pharmacol. 1999;58:605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley AM, Bonham Carter SM, Reveley MA, Sandler M. A genetic study of platelet phenolsulphotransferase activity in normal and schizophrenic twins. J. Psychiatr. Res. 1982;17:303–307. doi: 10.1016/0022-3956(82)90009-7. [DOI] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram RS, Szumlanski C, Otterness D, van Loon JA, Weinshilboum RM. Human intestinal phenol sulfotransferase: assay conditions, activity levels and partial purification of the thermolabile form. Drug Metab. Dispos. 1989;17:255–264. [PubMed] [Google Scholar]
- Van Loon J, Weinshilboum RM. Human platelet phenol sulfotransferase: familial variation in thermal stability of the TS form. Biochem. Genet. 1984;22:997–1014. doi: 10.1007/BF00499627. [DOI] [PubMed] [Google Scholar]
- Vietri M, Pietrabissa A, Mosca F, Spisni R, Pacifici GM. Curcumin is a potent inhibitor of phenol sulfotransferase (SULT1A1) in human liver and extrahepatic tissues. Xenobiotica. 2003;33:357–363. doi: 10.1080/0049825031000065197. [DOI] [PubMed] [Google Scholar]
- Waring RH, Ayers S, Gescher AJ, Glatt HR, Meinl W, Jarratt P, Kirk CJ, Pettitt T, Rea D, Harris RM. Phytoestrogens and xenoestrogens: the contribution of diet and environment to endocrine disruption. J. Steroid Biochem. Mol. Biol. 2008;108:213–220. doi: 10.1016/j.jsbmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Zhao JH. 2LD, GENECOUNTING and HAP: computer programs for linkage disequilibrium analysis. Bioinformatics. 2004;20:1325–1326. doi: 10.1093/bioinformatics/bth071. [DOI] [PubMed] [Google Scholar]