Abstract
Understanding the functional landscape of the mammalian genome is the next big challenge of biomedical research. The completion of the first phases of the mouse and human genome projects, and expression analyses using microarray hybridization, generate critically important questions about the functional landscape and structure of the mammalian genome: how many genes, and of what type, are there; what kind of functional elements make up a properly functioning gene? One step in this process will be to create mutations in every identifiable mouse gene and analyze the resultant phenotypes. Transposons are being considered as tools to further initiatives to create a comprehensive resource of mutant mouse strains. Also, it may be possible to use transposons in true forward genetic screens in the mouse. The “Sleeping Beauty” (SB) transposon system is one such tool. Moreover, due to its tendency for local hopping, SB has been proposed as a method for regional saturation mutagenesis of the mouse genome. In this chapter, we review the tools and methods currently available to create mutant mice using in vivo, germline transposition in mice.
Keywords: Sleeping Beauty, transposon, mouse transgenesis, insertional mutagenesis, germline mutagenesis
1. Introduction
1.1. Transposable Elements
There are two major classes of transposable elements. Class I transposons, also called retrotransposons, are mobilized through an RNA intermediate in a “copy-and-paste” manner. Thus, the donor site remains intact during the retrotransposition process. The LINE1 class of transposable elements is representative of this group and is active in many mammalian species including the mouse and human (1). Class II transposons move without going through an RNA intermediate. Instead, these transposons are mobilized by a “cut-and-paste” transposition process. Characteristic of class II transposons is an encoded transposase gene required for the transposition process. These transposase proteins usually recognize sequences within inverted terminal repeats that flank the transposon DNA and which are required for transposition. One large family of class II transposons is composed of Tc1/mariner elements (2). Tc1/mariner family elements are flanked by inverted terminal repeats and contain an encoded transposase gene with a paired-like DNA-binding domain and “DDE” catalytic domain (2). This family of transposable elements has been a source of several transposon systems that can be used to engineer vertebrate genomes.
1.2. The Sleeping Beauty (SB) Transposon System
The “Sleeping Beauty” (SB) transposon system, derived from inactive Tc1/mariner family transposable elements is currently the most well-studied “cut-and-paste” transposon available for use as a general gene transfer and insertional mutagenesis tool. Two parts of the SB system must be supplied to a cell for transposition to occur – the transposon vector DNA and the enzyme that mobilizes the vector DNA, the transposase. The SB transposase gene, the first version of which is called SB10, was created by derivation of a functional enzyme gene from multiple disabled, mutant copies of Tc1/mariner family transposase genes. These Tc1/mariner transposase genes had been cloned from the genomes of various species of salmonid fish, where they lay dormant for at least 10 million years (3). In order for the SB transposase to catalyze transposition, a sequence must be flanked by special sequences called inverted repeat/direct repeat (IR/DR) elements. The left and right IR/DRs consist of very similar inverted terminal sequences but they are distinct functionally (4). Moreover each IR/DR contains an inner and an outer DR element (hence IR/DR). These DRs are the binding sites for the transposase protein. Transposon vectors that contain the original IR/DRs built from sequences from salmonid fish are designated “pT” vectors (3). However, sequence changes have been introduced into the IR/DRs that increase transposition rates somewhat. Transposon vectors that are based on this second generation of IR/DRs are designated “pT2” vectors (5). Also, new versions of the SB transposase protein with improved catalytic activity have been developed in which one or more amino acid substitutions are introduced (6, 7).
A variety of insertional mutagenesis applications have been reported for SB including germline mutagenesis in the mouse and zebrafish (8) and most recently somatic mutagenesis for cancer gene discovery in the mouse (9, 10). Several other transposable element systems have now been shown to be active in the mouse germline, including Minos (11) and PiggyBac (12, 13). However, germline mutagenesis using SB has been most thoroughly described in the literature (14–22). The lessons learned using SB might in large part apply to these other transposon systems. Therefore, in this chapter we will focus on the use of SB for germline mutagenesis.
This chapter will detail the general methods used to create germline transposon insertion mutations using SB in the laboratory mouse. It is worth pointing out that it has recently been shown that SB germline mutagenesis is also very efficient in the laboratory rat (23, 24). Hence, the methods in this chapter apply also to the rat. Included in the description are the transgenic lines that have been described and that might be made for such a purpose. In addition, special concerns for isolation and analysis of the most useful mutant alleles will be described. This is a complex process from the conceptual point of view, yet in practice is easy to carry out. Using transposons for germline mutagenesis requires no special cell culture technologies or the use of chemicals. In fact, the techniques are similar in concept to paths that are well-worn in other model genetic organisms such as Drosophila melanogaster (25). Because this technology utilizes random insertion of transposon vectors to create mutant alleles, the choice of the internal components of the vector is especially important. Many possible insertion mutations might be created and so all these are not discussed in depth in this chapter. Instead, the reader is referred to published reviews on insertional mutagenesis strategies for in-depth discussion of vector construction (26, 27).
1.3. Creating Transposon Insertion Mutations Using Sleeping Beauty
Transposon design
The first component needed to achieve trans-position using SB is a transgenic line of mice carrying the transposon vector to be used as the insertional mutagen. In order to create germline mutations using SB it is by far most efficient to use multi-copy transgene arrays carrying the transposon vector as donors for transposition. Single-copy transposon vectors transpose infrequently in the germline of mice, ranging from once in five gametes to less than once in 100 gametes (16, 20). In contrast, most multi-copy transgene arrays yield 0.5–3 new transposon insertions per gamete (14, 16, 18). This increased rate of transposition seems to be due to more than just an increase in the amount of substrate for transposition (18) and may be due to methylation of transposon DNA in a multi-copy array (28). Thus, we and others have used transposon vector transgenes produced using standard pronuclear injection into FVB/n or C57BL/6 J mice, although other strain backgrounds should also work.
The rate of germline transposition achieved in these experiments is critical. This rate determines the number of new insertion mutations in genes that can be induced per offspring. SB transposon vectors have been shown to insert into new sequences at TA dinucleotides (3). Although the sequences immediately adjacent to the TA dinucleotide do influence the likelihood of insertion, on a genomic scale SB insertion is essentially random (29). The one exception to this randomness is the tendency of SB transposon vectors to insert near the donor locus after transposition. This phenomenon is called local hopping (16, 22). Local hopping, and associated genomic damage due to deletions, translocations, and other events will be discussed in more detail below.
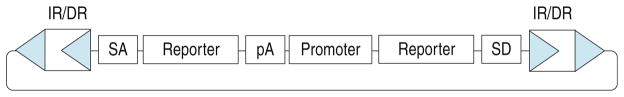
The transposon vector used for mutagenesis must be chosen with the desired downstream applications in mind. All described SB vectors, used for germline mutagenesis, include a so-called “5′ gene trap” including splice acceptors and polyadenylation sequences in one or both orientations so that genes can be disrupted upon insertion into an intron (22). In addition, some vectors include a reporter gene (GFP or LacZ) expressed from the chimeric mRNA generated by splicing of the endogenous transcript into the transposon vector (22). These reporters have generally been preceded by an internal ribosome entry site (IRES) so that the reporter can be translated and expressed regardless of the reading frame of the disrupted gene. In one report from our lab, the tTA transcription factor was used as a reporter so that a tet-regulated element (TRE) reporter could be used in conjunction with gene-trap alleles produced with this SB vector (18). A second component of some SB transposon vectors is a so-called “poly-A trap” immediately downstream of the 5′ gene trap. The poly-A trap is composed of an internal promoter, a reporter gene, and a splice donor. In the absence of nearby downstream exons and a polyadenylation signal, the poly-A trap produces an unspliced and non-polyadenylated RNA that is not exported from the nucleus efficiently and is unstable. If the transposon vector has landed in a transcription unit, in the same orientation as that transcription unit, then the RNA transcript initiated by the internal promoter splices to endogenous exons, is processed, and polyadenylated. Thus, the message is stabilized and the reporter is expressed. For SB vectors, the internal promoter that has been shown to be useful for this purpose is the CAGGS promoter (22), which is composed of sequences from the chicken beta actin and human cytomegalovirus immediate early promoters and can be expressed ubiquitously in some transgenic mice (30). By using the GFP gene as a the reporter for the poly-A trap the Takeda lab at Osaka University has succeeded in developing a system that can be used to identify generation 1 (G1) mice with a new transposon insertion into a gene just by visual inspection of neonatal pups for GFP expression (22). This system can thus be used to identify those offspring most likely to have an SB-induced gene mutation. A generalized SB transposon vector, which combines both 5′ and poly-A gene traps is shown in Fig. 20.1.
Fig. 20.1.
A generic SB transposon vector is shown. The left and right inverted terminal repeat/direct repeat (IR/DR) sequences must flank the transposon vector. Inside a 5′ gene trap consists of a splice acceptor (SA), internal ribosome entry site (IRES), reporter transgene, and polyadenylation (pA) site. This portion of the vector will interrupt the endogenous gene’s mRNA processing and result in expression of the reporter in the same spatiotemporal pattern as the disrupted gene. A second gene-trapping cassette, called the poly-A trap is included. The poly-A trap includes an internal promoter (CAGGS has been shown to work (22)), a reporter, and splice donor (SD), but no polyadenylation site. The poly-A trap reporter is expressed, and is expressed ubiquitously, only if the transposon lands in a transcription unit in the same orientation as that transcription unit. Thus, G1 mice likely to have a gene disruption can be identified by looking for expression of the poly-A trap reporter. GFP is useful as a reporter for this purpose (22).
Transposase transgenes
Three different sources of SB transposase have been used in published work on SB-induced germline insertion mutations. A CAGGS-SB10 transgene has been used by several labs to catalyze germline transposition (14, 21). Transgenes created using the CAGGS promoter are reported to be ubiquitously expressed (31). We have found that the CAGGS promoter is not ubiquitously expressed in all transgenic lines, but is expressed at very high levels in the developing germ cells of male mice in some transgenic lines (unpublished data). In one report, the mouse protamine 1 gene promoter (Prm1) has been used to drive SB10 expression and catalyze male germline transposition at good rates (20). Finally, a catalytically improved SB11 transposase cDNA under the control of the endogenous Rosa26 promoter (32) can be used to drive germline SB transposition at good rates (our unpublished data). In our experience both CAGGS-SB10 and Rosa26-SB11 transgenes catalyze SB transposon vectors in the male germline at similar rates, with a slight advantage to the CAGGS-SB10 transgene. The Prm1-SB10 transgene seems to work at rates similar to the CAGGS-SB10 transgene (20). At present, the CAGGS-SB10 transgene is the standard for germline mutagenesis. It should be mentioned that improved SB transposases, generated by amino acid substitutions, are being generated by several labs (7). These improved SB transposases promise to allow germline transposition rates much higher than three new insertions per gamete.
Generation of doubly transgenic “seed males” for SB mutagenesis
Once suitable transposon and transposase transgenic lines have been selected the next step is to breed mice together to obtain males that carry both transgenes. These doubly transgenic males are referred to as “seed males” in the literature because they are the source of sperm carrying new SB transposon vector insertions due to ongoing transposition in the developing sperm of these mice (18, 22). The seed males are bred to wild-type females to generate G1 offspring carrying new transposon insertions. The general outline of this procedure is shown in Fig. 20.2. We have found that seed males carrying identical transgenes can vary in the rate of observed germline transposition (18). A single male may vary in germline transposition rate during its lifetime as well (unpublished observations). For this reason, it is advised that multiple seed males should be generated and test litters generated from each male should be screened by Southern blot to identify those males with the highest germline transposition rate (18). Any one male is estimated to produce among its sperm at least 10,000 different transposon insertion mutations (22).
Fig. 20.2.
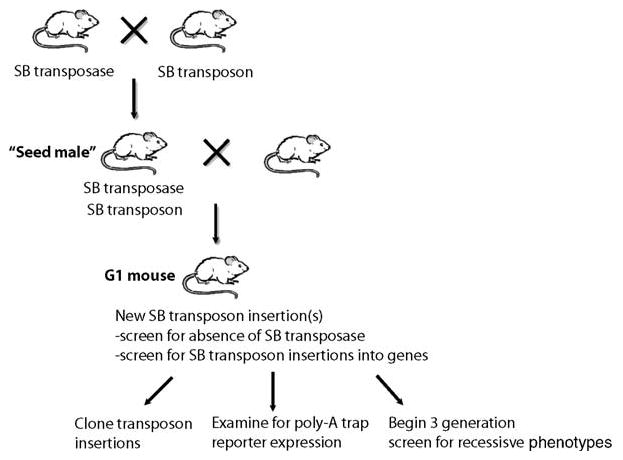
Outline of the crosses and steps used to create and analyze SB transposon insertion mutations. Doubly transgenic seed males, carrying both the transposase and transposon transgenes are generated. These males are bred to wild-type females to generate G1 offspring. The G1 offspring are screened for the absence of the SB transposase transgene and the presence of gene mutations by (1) cloning transposon insertions, (2) expression of 5′ or poly-A trap reporters, or (3) entering the G1 mice into a three-generation screen for recessive mutant phenotypes.
The analysis of G1 offspring including cloning and sequence analysis of transposon insertion sites
In general, the G1 generated from crossing a seed male to a wild-type female are screened for the presence of new transposon insertions into genes by any of several methods. If a CAGGS-GFP-SD poly-A trap was included in the vector, then GFP+ offspring are sought (22). If a visible marker was not included, then new transposon insertions can be sought via Southern blotting and/or cloned using inverse polymerase chain reaction (PCR) or linker-mediated PCR (16). An analysis of the transposon insertion site can be done, by comparison to the draft mouse genome sequence (http://www.ensembl.org) to determine if the insertion is likely to have impaired gene function. Finally, in one project from my own laboratory, we have bred G1 mice to create G2 mice, which were then intercrossed to determine if a mutant phenotype could be uncovered in mice homozygous for an SB mutagenized chromosome (17). It should be emphasized that any G1 mouse may harbor multiple, independent SB transposon insertion mutations. As mentioned above, roughly 50%–80% of these insertions will occur on the same chromosome as the donor locus, with many being within 10–20 Mbp of the donor locus (17). Thus, several SB transposon insertions may be linked to each other. The tendency of SB for local hopping has been proposed as a method for achieving regional saturation mutagenesis in the mouse (19). However, we discovered that about 40% of all G1 mice carry a deletion, inversion, or translocation of sequences adjacent to the donor locus (17). These chromosomal rearrangements affect hundreds of kilobase pairs and most often are deletions extending from one end of the donor locus. Since these deletions can remove essential genes, it means that the true homozygous phenotype of linked SB transposon insertions cannot be determined unless and until said insertion has been separated from the donor locus by meiotic recombination. Therefore, it is imperative to both identify all the SB insertions present within a G1 animal and separate those of interest from all other transposon insertions and the donor locus. The inheritance of individual transposon insertions can be followed by Southern blotting. All the new transposon insertions can be cloned using PCR-based strategies.
In order to process a large number of tail genomic DNAs and to approach saturation for insertion site recovery, we use PCR-based methods for amplifying insertion sites. Linker-mediated PCR is the most common technique currently used to clone transposon integration sites from mice carrying new SB transposon insertion sites (14, 16, 17). In this method, specially designed linkers are ligated onto restricted tumor genomic DNA, then subjected to two rounds of PCR using transposable element-specific and linker-specific primers before cloning into a plasmid vector for sequencing. The use of linker-mediated PCR amplification and cloning of transposon insertion sites is described in detail in another methods chapter in this series (Collier and Largaespada, Methods in Molecular Biology).
After G1 mice have been obtained and the transposon insertions of interest have been cloned and sequenced, the insertion mutations should be separated from other transposons by meiotic recombination (16). It is relatively simple to design primers useful for three-primer PCR reactions that can distinguish the unmodified, wild-type insertion site from the insertion allele (16). This process can be used to genotype mice and determine whether they are homozygous wild-type, heterozygous for the insertion mutation, or homozygous insertion mutants.
2. Materials
2.1. Transposon Transgenic Lines
Once an SB transposon vector has been generated, it should be used to create transgenic mice by standard pronuclear injection. The reader is referred to previous publications on this technology (33). Several SB transposon vectors have been described in the literature including 5′ gene and poly-A traps (14, 18, 22).
2.2. Transposase Transgenic Line
-
CAGGS-SB10/+ mice (14). These mice are available via the Mouse Models of Human Cancer Consortium (http://mouse.ncifcrf.gov/).
or
-
Prm1-SB10/+ mice (20)
or
Rosa26-SB11 mice (10).
3. Methods
3.1. Generation of Doubly Transgenic “Seed Mice”
A transposon vector should be created using recombinant DNA technology. Typically, gene-trapping components would include, at a minimum, splice acceptor and polyadenylation sequences to disrupt genes upon intronic insertion (see Notes 1 and 2).
Transposon transgenic mice are produced by standard pro-nuclear injection with linearized SB vector DNA (see Note 3). Alternatively, previously used SB transposon transgenic mice may be available (see above).
Several transgene positive founders should be characterized by Southern blotting for copy number. Several medium- and high-copy transposon transgenic lines should be tested for germline transposition rate (see Note 4).
Each transposon transgenic line is bred to SB transposase transgenic mice to create doubly transgenic male “seed mice” (see Note 5). Several of these seed mice should be tested for germline transposition by crossing to wild-type females and screening tail biopsy genomic DNA from the offspring by Southern blotting for new transposon insertions (see Note 4).
Once a transposon transgenic line has been chosen for further analysis as many G1 offspring carrying new transposon insertions as desired may be generated. Indeed, it would be theoretically possible to mutate most genes in the genome, by transposon insertion, if enough G1 mice are generated.
3.2. Generation and Analysis of G1 Offspring from Doubly Transgenic “Seed Mice”
G1 offspring can be screened for new transposon insertions using expression of the 5′ or poly-A trap reporter or by simply cloning the new transposon insertions from the G1 mouse. If the transposition rate is high enough it may be possible to simply screen G1 mice and derived G3 mice for new phenotypes without cloning the transposon insertions first. This “forward genetics” approach will probably require higher rates of germline transposition than are currently possible using SB. In any case, the SB transposon insertion mutations of interest must be identified by molecular cloning eventually.
SB transposon insertions are cloned by linker-mediated PCR (see Collier and Largaespada, Methods Mol Biol., 2008, for a description).
Preferably at the G1 generation, or at least the G2 generation, it is important to eliminate the SB transposase transgene from mutant mice (see Note 6). The presence of the SB transposase transgene could cause transposon remobilization, further transposon-induced insertion mutations or genomic rearrangements, most often deletions, at the transposon donor locus (17).
3.3. Propagation of Mutant Mice
Specific SB-induced transposon insertion mutations are propagated in mice simply by breeding.
Three-primer PCR reactions are used to distinguish the wild-type from the insertion mutant alleles (16).
Acknowledgments
This work was supported by grants R21 CA118600, R01 CA113636, and RO1 DA014764 from the National Institutes of Health (to DAL).
Footnotes
Careful consideration should be given to the SB transposon vector design before mice are created. One critical issue is how causality, linking a specific transposon insertion to a specific mutant phenotype, will be established. It is possible to revert SB transposon insertion mutations, and the attendant phenotype, by remobilization out of the affected gene (16). However, as mentioned above, SB transposon remobilization is inefficient using SB10 or SB11 transposase transgenes currently available. Thus, gene rescue or recreation of the mutation using embryonic stem cell technology could be considered for this purpose. However, these are technically demanding and slow approaches. Thus, it is advised that LoxP or Frt sites flank the gene-trapping portion of the transposon vector as in Geurts et al. (18). Thus, a Cre or Flp transgenic mouse could be used to “revert” the insertion mutation and for most intronic insertions a functional allele.
SB transposition is sensitive to the length of the transposon vector (34). While ~10-kbp transposons have been mobilized in the germline of mice (22), it is likely that smaller transposons will yield higher rates of transposition.
When the transposon vector DNA is used to create transgenic mice by pronuclear injection the vector should be first linearized or cut out of the plasmid in which it resides. However, it is important that adjacent plasmid sequences still linked to the transposon vector are retained in the DNA fragment that is injected. This plasmid DNA sequence should be incorporated into the transposon array that is generated for several reasons. Data from the Takeda lab suggest that plasmid sequences, and adjacent transposon sequences, may become methylated in the transgenic mouse which promotes SB transposition (28). Secondly, the plasmid sequences provide a good tag for the donor locus in genotyping reactions. Third, if the transposon sequences harbor one or more 6-bp restriction enzyme sites, then they can be used to digest linker-ligated genomic DNA during the transposon insertion site cloning protocol, thus eliminating donor transposons from those that are cloned in this reaction (16).
Several different transposon vector transgenic lines should be characterized for any vector. Those with the highest copy number tend to result in the most transposition activity when bred to SB transposase transgenic mice, but line-to-line variation is high due to position effects (18). Screening them for the typical transposition rate that is achieved in seed males is advised. New transposon insertions are detected by Southern blotting using a transposon vector-specific probe and cutting the genomic DNA with an enzyme that cuts once or not at all in the transposon vector. Thus, the transposon vectors that remain in the concatemers will result in a band of defined size while new transposon insertions will result in bands of a different size.
SB transposition is low in the female germline and so seed mice should be males (18).
When G1 animals are screened for new transposon insertions, it is helpful to also screen them for the presence of the transposase transgene. The transposase transgene will segregate independently of the transposon donor array if it resides on another chromosome. By eliminating G1 mice that carry the transposase transgene, one can ensure that no further SB transposon insertion events will occur and that the transposon insertion(s) of interest will remain stable in subsequent generations.
References
- 1.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 2.Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–32. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 3.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 4.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure–function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J Mol Biol. 2002;318:1221–35. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 5.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–17. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 6.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–47. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62:629–41. doi: 10.1007/s00018-004-4232-7. [DOI] [PubMed] [Google Scholar]
- 9.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 10.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 11.Drabek D, Zagoraiou L, deWit T, et al. Transposition of the Drosophila hydei Minos transposon in the mouse germ line. Genomics. 2003;81:108–11. doi: 10.1016/s0888-7543(02)00030-7. [DOI] [PubMed] [Google Scholar]
- 12.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39:922–30. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–8. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 15.Roberg-Perez K, Carlson CM, Largaespada DA. MTID: a database of Sleeping Beauty transposon insertions in mice. Nucleic Acids Res. 2003;31:78–81. doi: 10.1093/nar/gkg045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–56. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geurts AM, Collier LS, Geurts JL, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geurts AM, Wilber A, Carlson CM, et al. Conditional gene expression in the mouse using a Sleeping Beauty gene-trap transposon. BMC Biotechnol. 2006;6:30. doi: 10.1186/1472-6750-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keng VW, Yae K, Hayakawa T, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–9. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 20.Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci USA. 2001;98:6759–64. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horie K, Kuroiwa A, Ikawa M, et al. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc Natl Acad Sci USA. 2001;98:9191–6. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie K, Yusa K, Yae K, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitada K, Ishishita S, Tosaka K, et al. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–3. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 24.Lu B, Geurts AM, Poirier C, et al. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm Genome. 2007;18:338–46. doi: 10.1007/s00335-007-9025-5. [DOI] [PubMed] [Google Scholar]
- 25.Ryder E, Russell S. Transposable elements as tools for genomics and genetics in Drosophila. Brief Funct Genomic Proteomic. 2003;2:57–71. doi: 10.1093/bfgp/2.1.57. [DOI] [PubMed] [Google Scholar]
- 26.Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–68. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 27.Abuin A, Hansen GM, Zambrowicz B. Gene trap mutagenesis. Handb Exp Pharmacol. 2007:129–47. doi: 10.1007/978-3-540-35109-2_6. [DOI] [PubMed] [Google Scholar]
- 28.Yusa K, Takeda J, Horie K. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol Cell Biol. 2004;24:4004–18. doi: 10.1128/MCB.24.9.4004-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–52. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 30.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 31.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 32.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 33.Roths JB, Foxworth WB, McArthur MJ, Montgomery CA, Kier AB. Spontaneous and engineered mutant mice as models for experimental and comparative pathology: history, comparison, and developmental technology. Lab Anim Sci. 1999;49:12–34. [PubMed] [Google Scholar]
- 34.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–17. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]