To the Editor
Tumors release genomic DNA into the circulation of cancer patients following cellular necrosis and apoptosis. Isolation of the apoptotic fraction of plasma-circulating DNA can result to an enhanced detection of low-level mutations that can serve as tumor biomarkers (1). Because the amount of DNA circulating in the plasma of cancer patients is low, on the order of a few ng per ml blood, the number of genes that can be examined for tumor-specific alterations is limited, thus ultimately reducing biomarker sensitivity. We recently applied whole genome amplification of plasma-circulating DNA to increase the number of targets that can be analyzed from each sample, thus potentially increasing sensitivity (2). This approach yields highly-expanded DNA amounts for performing genetic screening, however there is no preferential enrichment of smaller-size DNA fragments. We describe a new method for whole-genome amplification of plasma-circulating DNA, based on Ligation-Mediated-PCR of Blunted DNA fragments (BLM-PCR), that results to preferential amplification of smaller size, apoptotic DNA fragments.
Plasma-circulating DNA was extracted from blood obtained from radiation therapy patients following consent and IRB approval. Within 2–3 h of collection, whole blood was centrifuged 2000g for 15–30 min, plasma was separated, and plasma-circulating-DNA was purified via QIAamp™ MinElute Virus Spin Kit (Qiagen) and quantified via Taqman-real-time-PCR. To test for Kras codon 12 mutations, a simplified version of the recently reported Fluorescent Amplicon Generation (FLAG) method (3) was used, that employs the highly thermostable PspGI enzyme to destroy wild-type alleles during PCR. Of 15 patients studied, three (#5, #6 and #15) were positive when testing the unamplified DNA from plasma (not shown). Tumor-derived DNA from patients #5, #6 and #15 also contained the plasma-identified Kras nucleotide-changes. Kras-positive samples #5, #6 and #15 plus two Kras-negative samples (#12, #13) were chosen for further study.
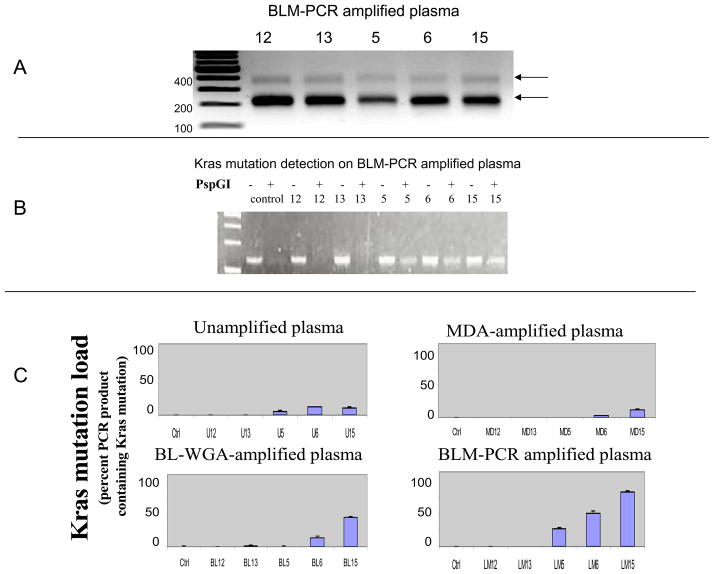
To apply BLM-PCR to these plasma-circulating-DNA samples, we generated blunt-ends on 2–5 ng plasma-circulating DNA using 0.6U T4 DNA polymerase at 12°C for 15 minutes in 5 μl ligase buffer supplemented with dNTP at a final concentration 100μmol/L. T4 DNA polymerase was then heat-inactivated. Double-stranded adaptors were prepared by annealing the following primers at 55°C: 5′-TTCCCTCGGATA-3′ and 5′-AGGCAACTGTGCTATCCGAGGGAA-3′. 5 μl of blunted DNA and 0.8 μl adaptors were then ligated via T4 DNA ligase. 60μl PCR reactions contained final reagent concentrations as follows: 1X GoTaq Flexi buffer, 1.5mM MgCL2, 2 mM each dNTP, 0.2 μM 24-mer primer and 6 μl adaptor-ligated product. The reaction was incubated at 72°C for 3 minutes followed by rapid cooling on ice. PCR-cycling was: 72 °C 5 min; 95°C, 2 min; (95°C, 15 sec; 72°C 15 sec) × 25 cycles, using 1.3U GoTaq Flexi DNA polymerase (Promega). When the BLM-PCR product was examined via gel-electrophoresis, a pattern of discrete DNA sizes of ~200 and ~400 bp was observed uniformly from all samples (Figure 1A). Plasma-circulating DNA consists of a mix of small fragments consistent with apoptotic DNA the size of mono-or di-nucleosomes and of large DNA fragments (1,4,5). As we demonstrated using larger size DNA, the conditions applied for BLM-PCR preclude amplification of DNA in excess of a few hundred base pairs. A similar ladder-like pattern was also observed when plasma-circulating DNA from healthy volunteers was amplified via BLM-PCR (not shown). When BLM-PCR samples were tested for Kras mutations, we found absence of Kras mutations for samples #12 and #13 (no PCR product in presence of PspGI) and clear presence of mutations for #5, #6 and #15 (Figure 1B). Two additional methods of whole-genome-amplification were also applied to the five plasma-circulating-DNA samples, BL-WGA (that amplifies all DNA fragment-sizes) and MDA (that amplifies only the large DNA fragment-sizes) (2) and Kras mutation-detection was repeated. The Kras mutation load (fraction of DNA product containing Kras mutations), was semi-quantitatively estimated by measuring the fraction of endpoint PCR product surviving PspGI digestion. For this purpose, samples were amplified either in the presence or in the absence of PspGI and relative PCR product was quantified via dHPLC. Following BLM-PCR, the three samples that were Kras-mutation positive demonstrated an increased Kras mutation load relative to other methods of amplification or relative to unamplified plasma-circulating DNA indicating an enhanced ability to detect Kras mutations (Figure 1C).
Figure 1.
Panel A. Blunt-ended ligation-mediated PCR of plasma-circulating DNA (BLM-PCR). This 3 step procedure involves (i) blunting of the ends of double-stranded plasma-circulating DNA using T4 DNA polymerase, (ii) ligation of blunt-ended adaptors and (iii) PCR-amplification. The DNA amplicons resulting from amplification of DNA from samples #12, 13, 5, 6, and 15 is indicative of a DNA-ladder-like pattern consistent with the amplification of mono- and di-nucleosomal DNA. Panel B. Detection of Kras mutations using the FLAG assay on BLM-PCR-amplified plasma-circulating DNA from patients #12, 13, 5, 6 and 15, using gel electrophoresis. All samples generate PCR product in the absence (−) of PspGI enzyme, however only samples with Kras mutations generate products in the presence (+) of PspGI. Samples #5, 6 and 15 present a substantial amount of residual PCR product in the presence of PspGI. Normal human male DNA was used as a negative control. Panel C. Comparison of Kras mutation load (percent PCR product surviving PspGI digestion) in plasma-circulating DNA amplified using 3 different whole genome amplficiation methods, and in unamplified DNA. The Kras mutation load in patient #5 is below detection level following MDA (that amplifies large DNA fragments) or BL-WGA (that amplifies all DNA fragments) whole genome amplification. BLM-PCR-amplified DNA results to a significant increase in the Kras mutation load on all 3 samples that are Kras-positive in unamplified plasma-circulating DNA (#5, 6 and 15). Normal human male DNA was used as a negative control. The histograms depict the average and standard deviation of 3 independent experiments, starting from unamplified plasma each time.
In summary, BLM-PCR provides selective amplification of apoptotic DNA and potentially a method to improve identification of rare alleles in plasma-circulating DNA while also providing ample DNA for testing an almost unlimited number of biomarkers for monitoring cancer patients. This technique is anticipated to be equally applicable to other applications such as prenatal diagnosis.
Acknowledgments
GRANTS/FINANCIAL SUPPORT
This work was supported in part by NIH grants CA-115439, CA-111994, by NIH training grant 5 T32 CA09078 (JL), and by the Joint Center for Radiation Therapy Foundation.
ACKNOWLEDEGMENTS
None
Footnotes
“This is an un-copyedited authored manuscript copyrighted by The American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article will be made available at http://www.clinchem.org 12 months after its publication in Clinical Chemistry.”
FINANCIAL DISCLOSURES
None of the authors on this manuscript has financial interest/conflicts to disclose
References
- 1.Wang M, Block TM, Steel L, Brenner DE, Su YH. Preferential isolation of fragmented DNA enhances the detection of circulating mutated k-ras DNA. Clin Chem. 2004;50:211–213. doi: 10.1373/clinchem.2003.026914. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Harris L, Mamon H, Kulke M, Liu W, Zhu P, Makrigiorgos GM. Whole genome amplification of plasma-circulating DNA enables expanded screening for allelic imbalance in plasma. Journal of Molecular Diagnostics. 2006;8:22–30. doi: 10.2353/jmoldx.2006.050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amicarelli G, Shehi E, Makrigiorgos GM, Adlerstein D. FLAG assay as a novel method for real-time signal generation during PCR: application to detection and genotyping of KRAS codon 12 mutations. Nucleic Acids Res. 2007;35:e131. doi: 10.1093/nar/gkm809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 5.Li Y, Zimmermann B, Rusterholz C, Kang A, Holzgreve W, Hahn S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem. 2004;50:1002–1011. doi: 10.1373/clinchem.2003.029835. [DOI] [PubMed] [Google Scholar]