Abstract
The primary event in the movement of a migrating eukaryotic cell is the extension of cytoplasmic sheets termed lamellipodia composed of networks of actin filaments. Lamellipodia networks are thought to arise through the branching of new filaments from the sides of old filaments, producing a dendritic array. Recent studies by electron tomography have revealed the three dimensional organization of lamellipodia and show, contrary to previous evidence, that actin filaments do not form dendritic arrays in vivo. These findings signal a reconsideration of the structural basis of protrusion and about the roles of the different actin nucleating and elongating complexes involved in the process.
Introduction
Migrating cells use actin filaments to push at the front, by polymerization and to pull at the rear, by forming contractile assemblies with myosin. Both processes are maintained by a continuous remodeling of the actin cytoskeleton, involving the cycling of actin between protruding and contracting domains [1,2]. Pushing takes place in lamellipodia and filopodia, cytoplasmic sheets and rods composed of networks and bundles of actin filaments, respectively. The last decade has witnessed a considerable advance in the characterization of the molecular machineries that regulate the turnover of actin and, in particular about the factors that nucleate and drive the polymerization process, with the result that propulsion by actin can now be reproduced in vitro [3]. Inevitably, many molecular complexes are involved and more are coming to light [4,5]. To understand how these various actin modulators collaborate in the cell, to generate and regulate protrusions, we need to define the biochemistry of their interactions, as well as the structural and mechanical properties of the actin assemblies that push.
Electron microscopy has been applied to studies of lamellipodia structure, using different types of preparation procedures [6], mostly with cells initially de-membranated with detergent, to reveal the underlying cytoskeleton. Using subsequent sample processing for electron microscopy that involved critical point drying and coating with platinum, Svitkina and colleagues [7] produced high contrast images of lamellipodia in fish keratocytes, a model cell type exhibiting rapid gliding movement. In addition to confirming the presence of diagonal actin arrays in the protruding regions of the keratocyte lamellipodium [8], new features of filament organization were described, in particular “T-junctions” or branches of actin filaments. The branches were particularly prominent in a zone around 1 μm wide at the lamellipodium front, where there were also many free filament ends producing a brush-like appearance [7]. This report shortly preceded findings describing the branching of actin filaments in vitro by the then newly characterized Arp2/3 complex [9].
The heptameric Arp2/3 complex [10] is a major player in lamellipodia formation that localizes to lamellipodia [11], and cycles with actin, from the tip to the base in a treadmilling mode [12]. When mixed with actin in vitro the Arp2/3 complex is a poor nucleator of actin polymerization [9], but the process is enhanced considerably by including a nucleation promoting factor of the WASP/WAVE family [13], normally the active C-terminal part (the VCA domain) and seeds of F-actin [10,14]. The in vitro nucleation process can be observed directly by TIRF (total internal reflection fluorescence) microscopy using fluorescent actin [15], whereby new filaments are seen to grow from the sides of pre-existing ones, as branches. The structure of the Arp2/3 complex has been solved at atomic resolution [16], and electron tomography has been applied to map the positions of the Arp2/3 complex subunits at the junction of actin branches formed in vitro [17]. Antibodies to the Arp2/3 complex were also found to decorate the “brush zone” in keratocytes [18]. Taken together, the observation of T-junctions in keratocyte lamellipodia, and the branching of actin by Arp2/3 complex in vitro, lead to the current dendritic nucleation/array treadmilling model of lamellipodia protrusion [19] (Box 1).
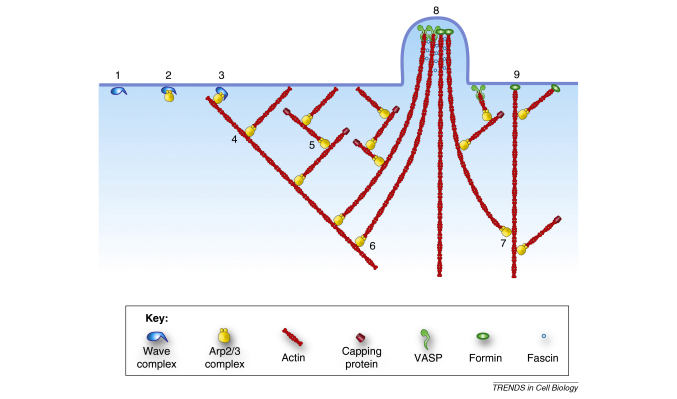
Box 1. The dendritic nucleation/array treadmilling model of lamellipodia protrusion.
The essential features of the model are depicted (Figure I) as originally proposed [19], with subsequent modifications to include filopodia formation from filaments generated in lamellipodia [47,57]. For simplicity, aspects of filament depolymerization at the rear of the lamellipodium, and recycling of actin monomer to the front are omitted. Steps 1-5 illustrate the sequence of events foreseen in the basic model: (1) Recruitment of the Scar/WAVE complex to the membrane via signal transduction pathways involving Rac and PIP2; (2) association of the Arp2/3 complex with the WAVE complex; (3) co-activation of the Arp2/3 complex by WAVE and simultaneous binding of Arp2/3 to the side of a “mother filament”; (4) nucleation and elongation of an actin filament branch with the free, polymerizing plus end pushing the membrane; (5) filament branches serve as “mother filaments” for nucleating new branches before being capped by capping protein. Steps 6-8 illustrate the proposed transitions of lamellipodia filaments into filopodia. In this scheme, filament branches originally nucleated on the WAVE/Arp2/3 complex (6,7) become associated at their plus ends with an elongation factor (e.g. Ena/VASP or a formin). The ends of the elongating filaments cluster, cross-link and generate a filopodium (8). It has been suggested that a formin could provide the mother filament to initiate the branched array (9) [47].
The validity of the dendritic nucleation model rests on the assumption that the electron microscope images of cytoskeletons, prepared by the critical point drying procedure, faithfully reflect the structure of lamellipodia actin networks. Advances in electron microscopy now allow structural analysis of the thin peripheral regions of cells vitreously frozen from the living state, allaying concerns about possible distortions introduced by conventional steps of sample preparation, such as chemical fixation and dehydration. Recent results obtained using this approach, combined with electron tomography, show that actin filaments in lamellipodia do not form dendritic arrays [20] and, we suggest, call for a re-evaluation of the structural basis of lamellipodia protrusion. The possible relevance of actin branching to other forms of movement involving actin is not excluded by these data.
Snags with branches in lamellipodia
A number of features of the dendritic nucleation model deserve closer inspection. First, an intuitively attractive feature of the model was that filament branching, with an inherent branch angle of 70 degrees appeared to explain the origin of the criss-cross network of actin filaments in lamellipodia seen by electron microscopy [21]. However, it is only in rapidly protruding or treadmilling regions of lamellipodia, like those found at the front of keratocytes,that actin filaments display a close to bimodal angular distribution [8,22]. In other situations, the angles that actin filaments subtend to the front edge of lamellipodia vary over a wide range, according to the rate of protrusion [1], inconsistent with a constant branch angle of 70 degrees. Second, the dendritic nucleation model relies on the pre-existence of “mother filaments” positioned close to the cell edge, onto which the Arp2/3 complex can dock and nucleate a branch. The origin of such mother filaments remains enigmatic [23], although it has been suggested that they could arise through the local severing by cofilin of filaments pre-existing close to the plasmalemma [24]. The suppression of cofilin activity does not, however inhibit the formation of lamellipodia [25]. Third, in the dendritic nucleation model [19], actin filaments formed on the sides of other filaments are thought to polymerize transiently before being capped at their growing ends by capping protein. It has been suggested that this produces short, stiff, uncapped filaments more suitable for pushing than longer, flexible filaments [19]. But this results in a large proportion of capped filaments unproductive in pushing, with the pushing filaments exerting force on the sides of the capped filaments in a mechanically unfavorable conformation (Box 1). Actin filament capping has also been perceived as a means of locally increasing the concentration of free actin monomers available for the growth of uncapped filaments, that can then grow faster [26], but this idea has recently been contested [27]. Capping protein is evidently important for lamellipodia formation, since its suppression causes the loss of lamellipodia in favour of filopodia [28], but its mechanism of action in vivo remains to be defined.
Tales from electron tomography
While many groups have focused on characterizing new actin modulators and their activities in vitro (reviewed in [4,5,10,29,30]), notably few have been concerned with determining the structural organization of actin arrays in lamellipodia by electron microscopy. As already indicated, a dendritic-like array of actin filaments is observed in lamellipodia of cytoskeletons prepared for conventional transmission electron microscopy by the critical point drying technique [18]. A similar impression of reticular networks of actin filaments in lamellipodia is apparent in images obtained after freeze-drying and platinum replication [31,32]. However, if cytoskeletons are instead embedded in an aqueous heavy metal salt, by negative staining, actin filaments appear more linear, and branching is not obvious [1]. Actin networks are fragile, and minor distortions of filaments, incurred through drying procedures, or uncontrolled ice crystal formation are enough to lead to misinterpretations of filament-filament interactions. Aside from the question of possible artifacts introduced by one or another technique [6], the lamellipodium is a three dimensional structure. It is therefore difficult to establish, from two-dimensional projections obtained by conventional electron microscopy, whether apparent filament intersections correspond to branch points or overlaps.
Advances in electron microscope technology have recently opened the way to electron tomography, also in combination with rapid freezing to avoid chemical fixation [33–35]. Early results of 3D imaging by this technique, with Dictyostelium amoeba, showed that actin filaments could be resolved in frozen cells [36], but the motile activity of the regions imaged was unknown. Electron tomography can be applied to those parts of cells thin enough to allow penetration of the electron beam. Lamellipodia, which vary from 0.1-0.3 μm in thickness, fall within this range, and tomograms of lamellipodia in cells frozen live and imaged in vitreous ice have been acquired recently [20]. Tracking of actin filaments through the tomograms showed that the lamellipodium is composed mainly of overlapping actin filaments. The same result was obtained for cytoskeletons dried in negative stain, which retain a three dimensional organization, albeit with a collapse to around 50% of the thickness of frozen preparations. These recent findings, including data from four cell types [20], indicate that the Arp2/3 complex nucleates un-branched arrays of actin in lamellipodia (Figure 1).
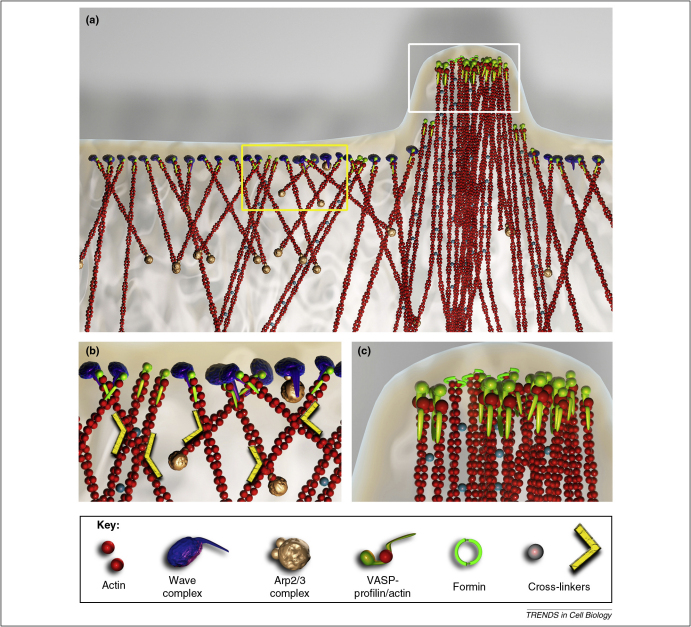
Figure 1.
Proposed tethered nucleation/elongation model of actin network formation in lamellipodia and transitions to filopodia. (a) Based on the results from electron tomography [20], actin filaments in lamellipodia form un-branched networks that include cross-linked filament pairs that can serve as potential precursors of filopodia. (b) Enlargement of the area boxed in yellow in A. In this hypothetical scheme, actin filaments are nucleated via the interaction of the Arp2/3 complex with the activated WAVE complex on the membrane. The Arp2/3 complex remains at the pointed end of the filament and treadmills with actin. The filament plus end elongates while being tethered to WAVE or to proximal Ena/VASP oligomers after transfer from WAVE. Filaments elongating on VASP can associate in pairs that are stabilized by a short actin cross-linker, like fascin. Long cross-linkers, such as filamin, stabilize the actin network. (c) Enlargement of the area boxed in white in A. Filopodia form via the clustering and bundling of actin filament pairs. At the filopodia tip, Ena/VASP proteins and formins cooperate in filament elongation. The inventory of proteins at lamellipodia and filopodia tips is far from complete; aside from missing adaptors, formins may also be involved in the generation of filament pairs in lamellipodia [20] and one of the WAVE isoforms, WAVE 2, is found at filopodia tips [58].
Setting up the network
If the Arp2/3 complex does not nucleate branched arrays of actin filaments in lamellipodia, the question arises as to how the actin network is established. The tomography data show single filaments abutting the lamellipodium tip, suggesting that filaments are nucleated individually on the membrane at different angles. However, we need more information about the early stages of lamellipodia induction to settle the question of network initiation. Theoretical treatments of the force velocity relationships of actin filaments in lamellipodia predict that, in established lamellipodia, filaments orient towards a regular bimodal angular distribution at a regular or high velocity of protrusion, independent of whether the filaments are branched or not [37,38]. During persistent protrusion, a condition exemplified by the fish keratocyte, the angular distribution of filaments may thus equilibrate around a value for which the protrusion rate is optimized, according to the constraints imposed by simultaneously pushing and inserting actin monomers between the filament plus end and the membrane. For filaments in filopodia, which extend at right angles to the membrane, other rules presumably apply. The question of whether or not unconventional myosins play a supporting role in pushing the plasmalemma, to facilitate actin monomer addition, remains an open possibility that has yet to be tested.
Different classes of actin cross-linkers evidently contribute to stabilizing the architecture of lamellipodia (Figure 1). The potent actin cross-linker, filamin, is present in lamellipodia [39,40], and cells depleted of filamin exhibit unstable lamellipodia and the tendency to bleb, pointing to a significant role of filamin in structuring actin networks [40]. Cross-linkers are probably engaged very early during lamellipodia formation, but, like fascin [41,42], may exchange rapidly to facilitate network plasticity. Actin cross-linkers, including fascin, probably contribute to the formation of filopodia through the generation of actin filament pairs recently identified as precursor building units of filopodia [20], which apparently derive from lamellipodia (Figure 1).
How are the actin filaments linked, if at all, to the membrane? In the “actoclampin” model [43], actin elongation complexes tether actin filaments to the membrane, and simultaneously allow insertion of actin monomers to drive filament growth. In vitro studies already support such a role for Ena/VASP proteins [44], which colocalize with WAVE at lamellipodia tips [44]. Currently unsettled is the question of how long a filament initiated by the association of Arp2/3 with WAVE on the membrane remains associated with the WAVE complex. If the VCA region of WAVE tethers actin filaments in the same manner as recently shown for N-WASP [45], the WAVE complex could serve an actoclampin-like role. Depletion of Ena/VASP proteins in fibroblasts leads to apparently shorter filaments in lamellipodia [46], consistent with a role of Ena/VASP proteins in the elongation of filaments, presumably primed through the WAVE/Arp2/3 complex pathway. Exogenously expressed formins also localize to lamellipodia [47,48], but the contribution of formin family members to filament generation in the network is currently speculative [20,47].
Pushing ahead
To develop a viable scheme of events at the advancing cell front, we must define the specific in vivo roles of the different nucleators, elongators and cross-linkers of actin filaments [5,49], as well as their modes of collaboration, in generating lamellipodia and filopodia. We also need to characterize the initial steps of lamellipodia formation, using suitable experimental models and conditions. To address these issues, correlated live cell imaging and electron tomography can be expected to play an important role by linking protein dynamics with cyto-architecture. The way is now open to manipulate living cells on the light microscope, for example by micro-injection of proteins or their functional domains, and to correlate the dynamic changes observed in lamellipodia and filopodia with changes in actin filament organization by electron tomography.
With the further evolution of super resolution light microscopy [50] we can expect the salient features of the actin cytoskeleton to become visible in more detail, perhaps in living cells, allowing the analysis of filament reorientations during movement, together with their dynamic interactions with associated proteins, with myosin filaments and other cytoskeletal elements. Electron tomography will be required to validate some of these imaging techniques and to provide high-resolution three-dimensional information not achievable by light microscopy. At the same time, new methods must be developed to allow the localization, by electron tomography, of actin-associated proteins, under conditions for which actin filament substructure and network superstructure is preserved.
Aside from lamellipodia protrusion, branched actin filament arrays have been implicated in endocytosis [51,52], the formation of adhesion structures termed podosomes [53,54], the propulsion of pathogens that invade the cytoplasm [55] and in vitro mimetic models of actin-based movement [56]. These latter phenomena depend on specific receptors, or N-WASP, rather than WAVE to activate the Arp2/3 complex [55], and require spatial geometries of actin filaments different from lamellipodia that could depend on branched interactions. Electron tomography will be necessary to clarify the organization of the pushing machinery in all these situations. There is still some way to go to unravel the different mechanisms by which actin polymerization is harnessed to push.
Figure I.
The dendritic nucleation model.
Acknowledgements
The author thanks Tibor Kulcsar for graphics and group members for helpful discussion. This work was supported by grants from the Austrian Science Research Fund (FWF project P21292-B09) and the Vienna Science Research and Technology Fund (WWTF).
References
- 1.Koestler S.A. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat. Cell Biol. 2008;10:306–313. doi: 10.1038/ncb1692. [DOI] [PubMed] [Google Scholar]
- 2.Nemethova M. Building the actin cytoskeleton: filopodia contribute to the construction of contractile bundles in the lamella. J. Cell Biol. 2008;180:1233–1244. doi: 10.1083/jcb.200709134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loisel T.P. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 4.Dominguez R. Actin filament nucleation and elongation factors — structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesarone M.A., Goode B.L. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small J.V. Unravelling the structure of the lamellipodium. J. Microsc. 2008;231:479–485. doi: 10.1111/j.1365-2818.2008.02060.x. [DOI] [PubMed] [Google Scholar]
- 7.Svitkina T.M. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small J.V. Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullins R.D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard T.D. Regulation of actin filament assembly by arp2/3 complex and formins. Annu. Rev. Biophys Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 11.Welch M.D. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai F.P. Arp2/3 complex interactions and actin network turnover in lamellipodia. Embo. J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takenawa T., Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 14.Machesky L.M. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amann K.J., Pollard T.D. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson R.C. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 17.Rouiller I. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svitkina T.M., Borisy G.G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 20.Urban E. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat. Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 21.Small J.V. The actin cytoskeleton. Electron Microsc. Rev. 1988;1:155–174. doi: 10.1016/s0892-0354(98)90010-7. [DOI] [PubMed] [Google Scholar]
- 22.Verkhovsky A.B. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol. Biol. Cell. 2003;14:4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard T.D., Cooper J.A. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DesMarais V. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- 25.Nishita M. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaloni D. Mechanism of actin-based motility. Science. 2001;292:1502–1506. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- 27.Akin O., Mullins R.D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mejillano M.R. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Insall R.H., Machesky L.M. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Bugyi, B., and Carlier, M.F. Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys 39, 449–470 [DOI] [PubMed]
- 31.Heuser J.E., Kirschner M.W. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J. Cell Biol. 1980;86:212–234. doi: 10.1083/jcb.86.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura F. Structural basis of filamin A functions. J. Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIntosh R. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Steven A.C., Aebi U. The next ice age: cryo-electron tomography of intact cells. Trends Cell Biol. 2003;13:107–110. doi: 10.1016/s0962-8924(03)00023-0. [DOI] [PubMed] [Google Scholar]
- 35.Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 2002;12:679–684. doi: 10.1016/s0959-440x(02)00378-0. [DOI] [PubMed] [Google Scholar]
- 36.Medalia O. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 37.Mogilner A., Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weichsel J., Schwarz U.S. Two competing orientation patterns explain experimentally observed anomalies in growing actin networks. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6304–6309. doi: 10.1073/pnas.0913730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langanger G. Ultrastructural localization of alpha-actinin and filamin in cultured cells with the immunogold staining (IGS) method. J. Cell Biol. 1984;99:1324–1334. doi: 10.1083/jcb.99.4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flanagan L.A. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 2001;155:511–517. doi: 10.1083/jcb.200105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa H. Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett. 2006;580:3223–3228. doi: 10.1016/j.febslet.2006.04.082. [DOI] [PubMed] [Google Scholar]
- 42.Vignjevic D. Role of fascin in filopodial protrusion. J. Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickinson R.B. Models for actin polymerization motors. J. Math. Biol. 2009;58:81–103. doi: 10.1007/s00285-008-0200-4. [DOI] [PubMed] [Google Scholar]
- 44.Breitsprecher D. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Co C. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bear J.E. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 47.Yang C. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Block J. Filopodia formation induced by active mDia2/Drf3. J. Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- 49.Small J.V. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 50.Schermelleh L. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perrais D., Merrifield C.J. Dynamics of endocytic vesicle creation. Dev. Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Kaksonen M. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 53.Mizutani K. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62:669–674. [PubMed] [Google Scholar]
- 54.Gimona M. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Goley E.D., Welch M.D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 56.Wiesner S. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J. Cell Biol. 2003;160:387–398. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svitkina T.M. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakagawa H. IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J. Cell Sci. 2003;116:2577–2583. doi: 10.1242/jcs.00462. [DOI] [PubMed] [Google Scholar]