Abstract
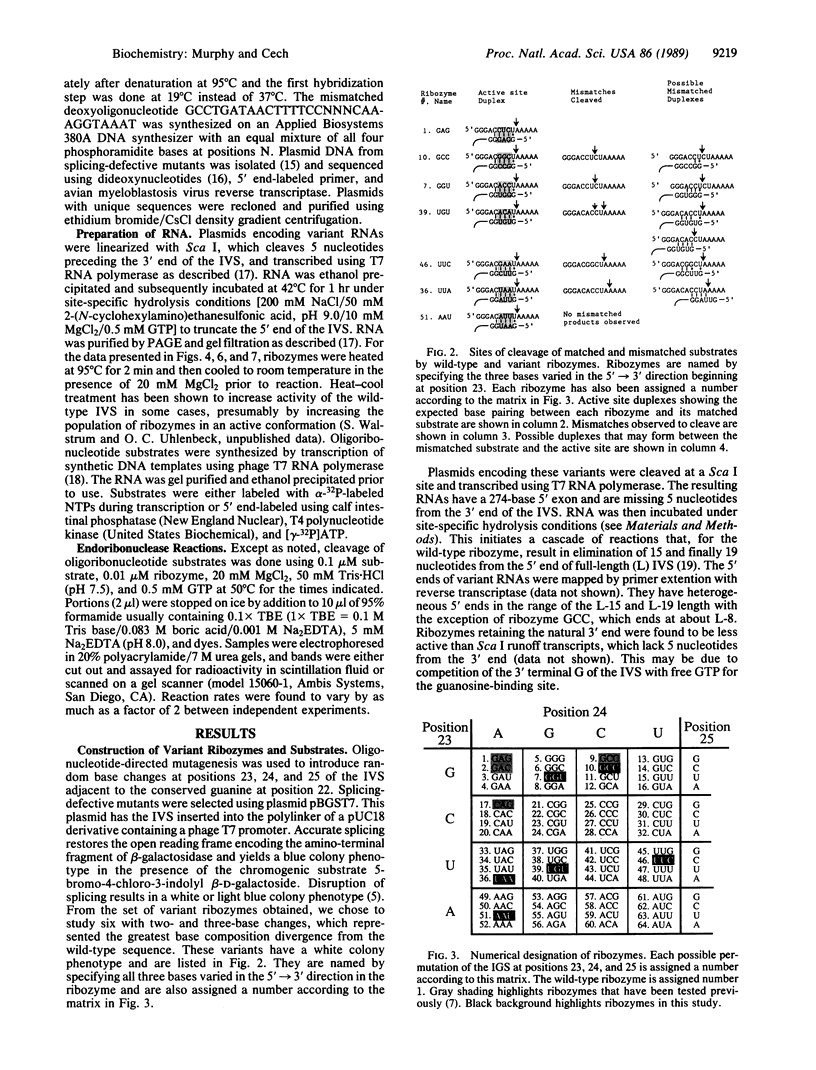
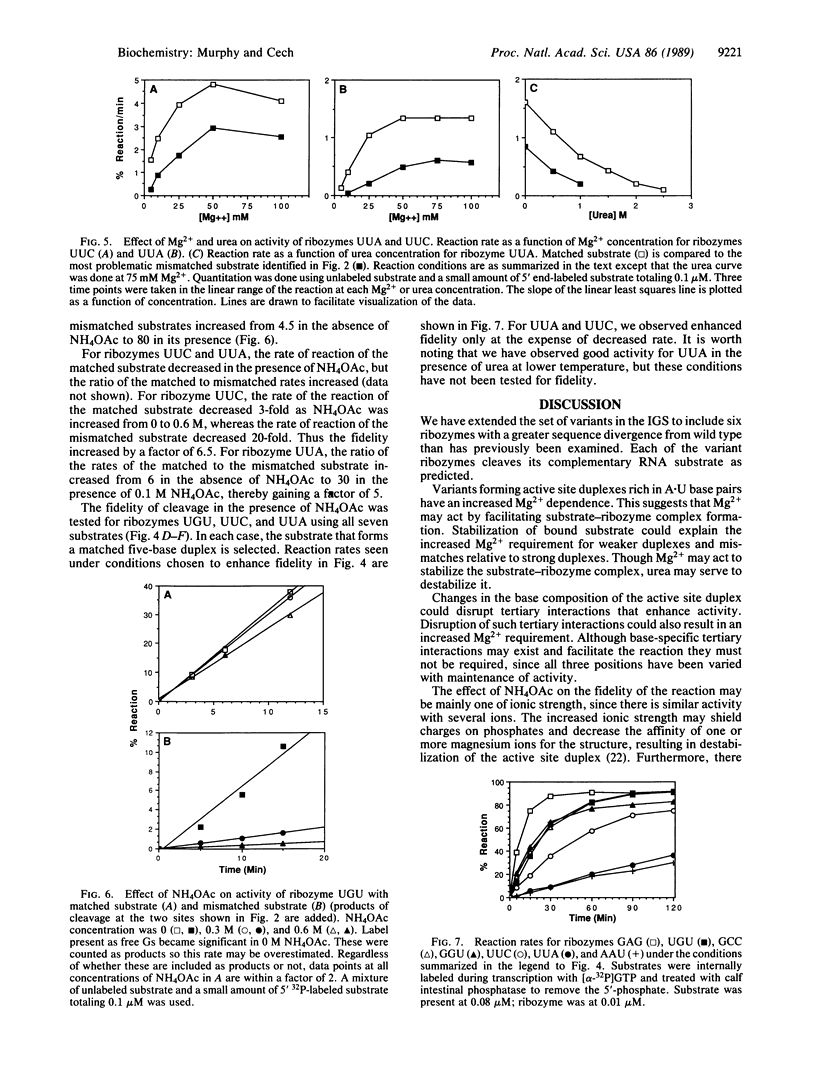
A shortened form of the intervening sequence of the self-splicing RNA from Tetrahymena thermophila catalyzes sequence-specific cleavage of RNA. Cleavage site selection involves a base-pairing interaction between the substrate RNA and a binding site within the intervening sequence. Single-base changes in this binding site were previously shown to alter substrate specificity in a predictable manner. To examine the generality with which substrate specificity can be altered, six variant catalytic RNAs (ribozymes) have been produced with two- or three-base changes in the active site. Each ribozyme cleaves its predicted substrate. The conditions required for good reactivity and for discrimination against cleavage at mismatched sites vary and were independently determined for each ribozyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barfod E. T., Cech T. R. The conserved U.G pair in the 5' splice site duplex of a group I intron is required in the first but not the second step of self-splicing. Mol Cell Biol. 1989 Sep;9(9):3657–3666. doi: 10.1128/mcb.9.9.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell. 1987 Sep 11;50(6):951–961. doi: 10.1016/0092-8674(87)90522-8. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Towner P. Internal guide sequence and reaction specificity of group I self-splicing introns. Cold Spring Harb Symp Quant Biol. 1987;52:165–171. doi: 10.1101/sqb.1987.052.01.021. [DOI] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry. 1989 Aug 22;28(17):6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989 Jun 13;28(12):4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Kay P. S., Inoue T. Catalysis of splicing-related reactions between dinucleotides by a ribozyme. 1987 May 28-Jun 3Nature. 327(6120):343–346. doi: 10.1038/327343a0. [DOI] [PubMed] [Google Scholar]
- Kay P. S., Menzel P., Inoue T. Two guanosine binding sites exist in group I self-splicing IVS RNAs. EMBO J. 1988 Nov;7(11):3531–3537. doi: 10.1002/j.1460-2075.1988.tb03229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N., Kierzek R., Turner D. H. Kinetics for reaction of a circularized intervening sequence with CU, UCU, CUCU, and CUCUCU: mechanistic implications from the dependence on temperature and on oligomer and Mg2+ concentrations. Biochemistry. 1988 Aug 23;27(17):6384–6392. doi: 10.1021/bi00417a029. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Tomka M., Kierzek R., Bevilacqua P. C., Turner D. H. Effects of substrate structure on the kinetics of circle opening reactions of the self-splicing intervening sequence from Tetrahymena thermophila: evidence for substrate and Mg2+ binding interactions. Nucleic Acids Res. 1989 Jan 11;17(1):355–371. doi: 10.1093/nar/17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Williams A. P., Longfellow C. E., Freier S. M., Kierzek R., Turner D. H. Laser temperature-jump, spectroscopic, and thermodynamic study of salt effects on duplex formation by dGCATGC. Biochemistry. 1989 May 16;28(10):4283–4291. doi: 10.1021/bi00436a025. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]