Abstract
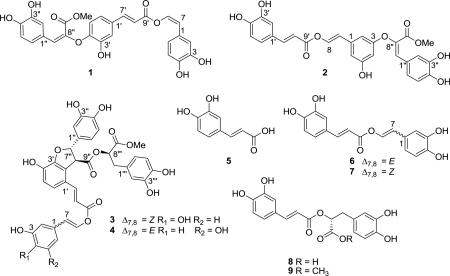
Bioassay-guided fractionation of an extract prepared from the fruits of Cordia sebestena has led to the isolation of sebestenoids A-D (1-4). The structures of these new phenylpropanoid esters were elucidated on the basis of extensive NMR experiments and mass spectroscopic measurements. Compounds 1-4 exhibited moderate inhibition of the aspartic protease BACE1.
Keywords: Cordia sebestena, Boraginaceae, Phenylpropaniods, BACE1
1. Introduction
Cordia sebestena (L.) (Boraginaceae Family) is commonly known as the Geiger tree. Hawaiians refer to the plant as Kou Haole though, which roughly translates to “foreign plant” (Abbott, 1992). Recent archaeological evidence indicates that the plant is actually indigenous to the islands (Burney et al., 2001). Regardless of its origin, the plant has a long history of use in Hawaiian culture. The plant's large dark green leaves have often been used to dye kappa, or wood cloth, that was used for both clothing and bedding. C. sebestena's dark orange flowers are typically used to make Leis. The plant is best known in the Hawaiian Island for its wood, which due to their lightweight, durable and easily workable nature, are used for many traditional items ranging from canoes to food vessels. The plant can grow up to 25 feet tall in tropical and sub-tropical areas where it is widely distributed due to its extensive use in landscaping. Despite its prevalence, C. sebestena's phytochemical or pharmacological properties have not been reported. Whether this species was used in traditional Hawaiian medicine is not clear, as there are vague references stating “many parts were used medicinally” (Krauss, 1993) although no specific diseases or preparations are mentioned and the plant is not included in more comprehensive texts on the subject (Chun and Hawaii, 1994; Chun and Kapunihana, 1998; Gutmanis, 1976; Kaaiakamanu and Chun, 2003; Kaitin, 2010).
As part of a program to examine Hawaii plants for bioactive components, we discovered that the EtOAc extract of the white fruit of C. sebestena showed activity against the aspartic protease BACE1, which is central to Alzheimer's disease etiology. Bioassay-guided fractionation of this extract has now led to the isolation four new active phenylpropanoids, given the trivial names sebesteniods A-D (1-4) that are described below. These new metabolites were accompanied by five known compounds, including: caffeic acid (5), netpetoidin A-B (6,7) (Arihara et al., 1975; Grayer et al., 2003), rosmarinic acid (8) and its methyl ester (9). In this paper, we describe the isolation, structure elucidation of these new compounds by spectroscopic methods along with an assessment of their bioactivity against BACE1.
2. Results and Discussion
2.1 Structure elucidation of the new compounds
Repeated orthogonal chromatographic separations of the crude extract led to isolation of the optically inactive compound 1 as an amorphous light yellow powder. Mass spectrometry analysis of this sample provided a [M + H]+ pseudomolecular ion at m/z 507.1288 that defined a molecular formula of C27H22O10. Solutions of this powder displayed a complex UV chromophore, which included allowed absorbances at 334 and 343 nm, that implied the presence of a highly conjugated system. This conclusion was supported by prominent vibrations attributable to carbonyls (1702 cm-1) and aromatic rings (1603 and 1504 cm–1) in the IR spectrum, in addition to those for hydroxyl groups (3403 cm–1). Considering the source phylum, the 17 degrees of unsaturation and the number of oxygens required by the molecular formula, 1 was likely a polyphenol.
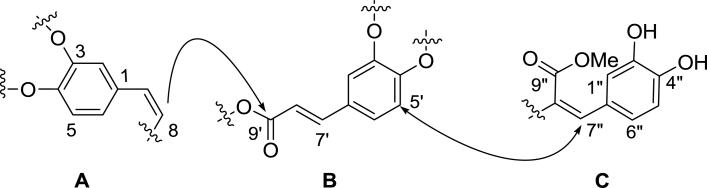
Analyses of the 1H (Table 1) and 2D NMR data (Table S1) established three fragments A-C (Figure 1) which confirmed the compound was a polyphenol derivative. Each fragment shared a similar catechol core as indicated by the three ABX systems observed in the 1H NMR spectrum, for example, fragment A was comprised of δH-5 6.73 (d, J = 8.2 Hz), δH-6 6.89 (dd, J = 8.2, 2.1 Hz), δH-2 7.28 (d, J = 2.1 Hz). On the basis of HMBC correlations observed in the 2D NMR data, two of the fragments, A and B, had disubstituted alkenes appended to aromatic cores, while C contained a trisubstituted olefin. The two carbonyls observed in the 13C NMR spectrum were substituents on these alkenes on the basis of HMBC correlations from H-7’ (B) or H-7” (C) to carbons at δC-9’ 165.4 and δC-9” 165.6, respectively. This latter carbonyl was clearly a methyl ester on the basis of a HMBC correlation from a downfield methyl singlet at δH 3.72 (C).
Table 1.
1H NMR Spectroscopic Data for Compounds 1-4 (500 MHz, δ ppm, J in Hz).
| Positionb | 1a | 2a | 3a | 4a |
|---|---|---|---|---|
| 2 | 7.28, d (2.1) | 6.69, d (1.5) | 7.27, d (2.0) | 6.67, d (1.7) |
| 4 | 6.83, d (1.5) | 6.81, d (1.7) | ||
| 5 | 6.73, d (8.2) | 6.75, d (8.2) | ||
| 6 | 6.89, dd (8.2, 2.1) | 6.70, br s | 6.92, dd (8.2, 2.0) | 6.68, brs |
| 7 | 5.63, d (7.2) | 6.36, d (12.8) | 5.61, d (7.3) | 6.35, d (12.8) |
| 8 | 7.22, d (7.2) | 7.78, d (12.8) | 7.21, d (7.3) | 7.76, d (12.8) |
| 2' | 7.27, d (2.0) | 7.23, d (2.0) | ||
| 5' | 6.76, d (8.2) | 6.74, d (8.4) | 6.87, d (8.3) | 6.84, d (8.4) |
| 6' | 7.04, dd (8.2, 2.0) | 7.00, dd (8.4, 2.0) | 7.30, d (8.3) | 7.22, d (8.4) |
| 7' | 7.75, d (15.8) | 7.69, d (15.8) | 7.80, d (15.9) | 7.70, d (15.9) |
| 8' | 6.55, d (15.8) | 6.42, d (15.8) | 6.44, d (15.9) | 6.29, d (15.9) |
| 2" | 7.24, d (2.0) | 7.26, d (2.0) | 6.75, d (2.0) | 6.73, d (2.1) |
| 5" | 6.73, d (8.4) | 6.72, d (8.4) | 6.74, d (8.2) | 6.75, d (8.2) |
| 6" | 7.08, dd (8.4, 2.0) | 7.07, dd (8.4, 2.0) | 6.63, dd (8.2, 2.0) | 6.62, dd (8.2, 2.1) |
| 7" | 7.33, s | 7.33, s | 5.81, d (4.4) | 5.80, d (4.6) |
| 8" | 4.42, d (4.4) | 4.38, d (4.6) | ||
| 2"' | 6.56, d (2.0) | 6.59, d (2.0) | ||
| 5"' | 6.56, d (8.1) | 6.62, d (8.1) | ||
| 6"' | 6.36, dd (8.1, 2.0) | 6.40, dd (8.1, 2.0) | ||
| 7"' | 2.84, dd (14.2, 9.2) | 2.91, dd (14.2, 9.2) | ||
| 2.96, dd (14.2, 4.5) | 3.02, dd (14.2, 4.0) | |||
| 8"' | 5.16, dd (9.2, 4.5) | 5.19, dd (9.2, 4.0) | ||
| OCH3 | 3.72, s | 3.72, s | 3.62, s | 3.67, s |
Data measured in MeOH-d4
Compounds numbered as in Murata et al. (2009).
Fig. 1.
Partial Fragments of 1 (A-C) along with selected HMBC (H→C) and ROESY (H↔H) correlations.
An atom inventory indicated the remaining oxygens were in the form of five hydroxy groups and either one or two ether linkages depending on whether C-9’ was an ester or a carboxylic acid. This latter possibility was excluded on the basis of the HMBC correlation observed from the vinyl proton H-8 (A) to the carbonyl carbon C-9’ (B), which linked these two units. Discriminating between the various possible combinations of ether linkages to C required detailed analyses of the ROESY spectrum. This spectrum displayed a cross-peak between H-5’ of fragment (B) and H-7” (C), which would only be observed if these two fragments were connected at the C-4’ oxygen of fragment B. The final planar structure of 1 was thus established as depicted after assignment of the olefin geometries on the basis of coupling constants (Δ7,8 cis, 3JH7,H8 = 7.2 Hz, Δ7’,8’ trans, 3JH7’,H8’ = 15.8) and ROESY analyses.
Sebestenoid B (2) was obtained as an amorphous light yellow powder, which had a molecular formula of C27H22O10 consistent with an isomer of 1. The NMR spectroscopic data of 2 were quite similar to those of 1, which allowed facile identification of fragments B and C originally found in 1. Differences were noted for resonances of fragment A that were attributed to changes in the substitution pattern of the aromatic ring and the configuration of the disubstituted alkene. Specifically, the ortho coupling (8.2 Hz) observed in 1 was missing in 2, replace instead with a smaller meta coupling (1.5 Hz) and corresponding upfield shift of the three aromatic protons in that moiety (δH-2 6.69, d, J = 1.5 Hz; δH-4 6.83, d, J = 1.5 Hz; δH-6 6.70, br s) indicative of 1,3,5 substitution pattern on the ring. Likewise, the vicinal coupling between the alkene protons was now consistent with a trans rather than cis system (1 JH-7/H-8= 7.2 Hz; 2 JH-7/H-8= 12.8 Hz). With these principal differences established, this 2-(3,5- dihydroxyphenyl)ethenyl fragment was connected to B via the same HMBC correlation observed for 1 thus defining the planar structure of 2.
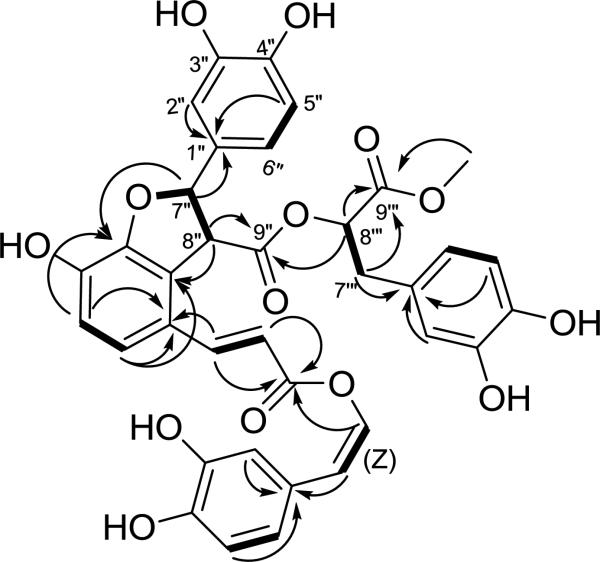
Compound 3 provided a pseudo-molecular ion peak at m/z 709.1532 [M + Na]+. In conjunction with the 13C NMR data, this established the molecular formula of 3 as C36H30O14 indicating 3 was larger than 1 or 2. A comparison of the spectroscopic data with the other compounds isolated from this extract identified the presence of fragment A (C-1 to C-8), as observed in 1, and the alpha-hydroxy ester subunit (C-1’” to C-9’”) found in the known rosmarinic acid derivative 9. This latter unit was also later confirmed by hydrolysis (vide infra). Analysis of the 2D NMR data established a catechol system, which had a sp3 hybridized carbon in the benzylic position rather than the typical sp2-hybridized carbon. This unit was expanded into a two carbon unit on the basis of a COSY correlation between this downfield proton and the H-8” methine. Beginning with these two resonances, analysis of HMBC correlations established that this two carbon unit was part of a larger dihydrobenzofuran ring system. Specifically, correlations observed between H-7”/C-3’, H-5’ to C-3’, H-8”/C-2’, and H-6’ to C-2’ were crucial for establishing the basic scaffold, while the additional correlations (Figure 2) allowed for the final structure of this compound to be deduced. An isomer 4 that differed both in the geometry of the C-7/C-8 enol ether and the substitution pattern on the aromatic ring was also identified from the same fraction and identified in a similar fashion.
Fig. 2.
Selected 1H-1H COSY (bold solid bars) and 1H-13C HMBC (H→C) correlations of 3.
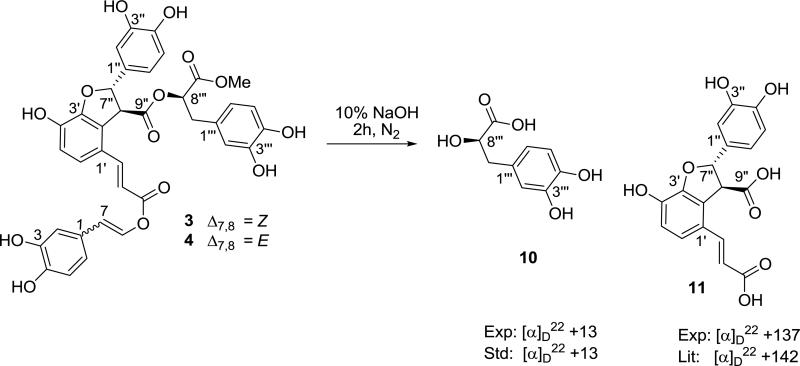
The stereochemistry of compound 3 was assigned using several techniques. The E and Z-configurations of the C-7’/C-8’ and C-7/C-8 olefins were elucidated on the basis of their characteristic 3JH,H values (3JH-7’,H-8’ = 15.9 Hz, 3JH-7,H-8 = 7.3 Hz) as was the trans relationship of the substituents on the dihydrofuran ring (3 3JH-7”,H-8”= 4.4 Hz, Lit: 3JHH= 4.9 Hz) (Wada 1992). To deduce the absolute configuration a mixture of 3 and 4 was hydrolyzed with a solution of sodium hydroxide under a N2 atmosphere (Fig. 3). After acid-exchange and reversed–phase chromatography 10 and prolithospermic acid (11) were isolated. A 8’”R configuration was established for 10, on the basis of comparison of the optical rotation with a standard prepared from hydrolysis of rosmarinic acid 8 (Fig. 3). The 7”S, 8”S configuration of 11 was deduced in a similar manner using optical rotation data previously reported (Wada 1992) (Murata et al., 2009). While epimerization of the cis-dihydrofuran derivative under basic conditions has been noted (Wada 1992), under our conditions the corresponding trans derivative is stable. The nearly identical magnitude of the vicinal proton-proton coupling values at the epimerizable stereogenic center (C-8”) before and after hydrolysis for this moiety support this conclusion (3 3JH-7”,H-8”= 4.4 Hz, 11 3JH-7”,H-8” = 5.1 Hz; if cis 3JH,H = 9.2 Hz (Wada 1992).
Fig.3.
Degradation of 3 and 4 to assign absolute configuration.
There are few reports describing the isolation of natural products possessing the (3,5-dihydroxyphenyl)ethenyl ethers (Nakanishi et al., 1990), making this a relatively distinct aspect of 2 and 4. In our hands, the 3,4-dihydroxyphenyl derivatives 1 and 3 were stable, while the corresponding 3,5-dihydroxyphenyl analogs 2 and 4 decomposed rapidly after isolation in pure form. Discoloration began almost immediately upon isolation, resulting in the formation of a complex mixture of products as determined by LC-MS with both higher and lower molecular weight products present. Mixtures of 3 and 4 were stable enough for prolonged storage in the freezer. Presumably, hydrogen bonding which is only possible in the 3,4-dihydroxyl isomer and a strongly electron-donating group para to the side chain in 1 and 3 both stabilize the enol ether, relative to 2 and 4. Consistent with this latter proposal, are the observations that electronegative substituents significantly increase the rate of vinyl ester hydrolysis (Euranto, 1977). Circumstantial evidence suggests a similar phenomenon may occur with the appropriately substituted caffeic acid derivatives as well. While the (3,4-dihydroxyphenyl)ethenyl derivative has been reported frequently (Grayer et al., 2003), there is only one report of the corresponding (3,5-dihydroxyphenyl) analog, and both are reported only detectable when extracted with organic solvent (Grayer et al., 2003) as they do not survive herbarium sample preparation of freeze-drying.
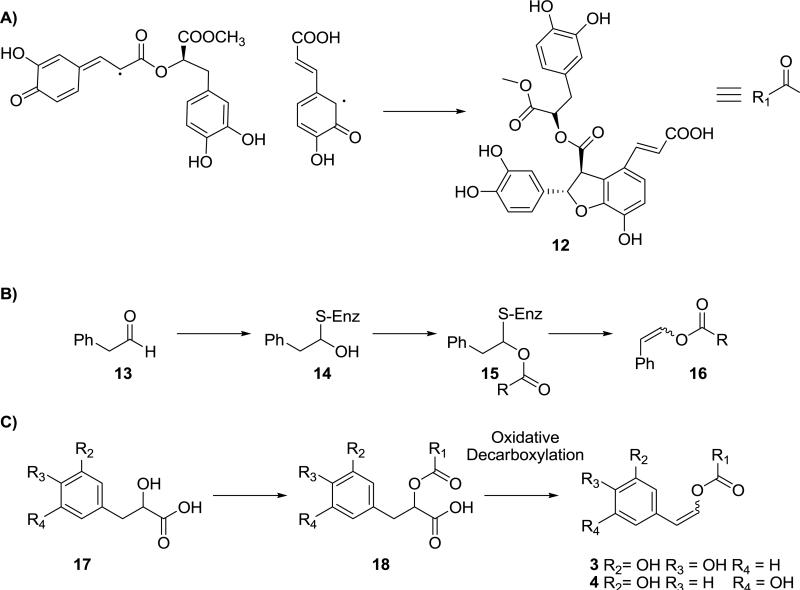
The structures of 3 and 4 raise an interesting question regarding their biosynthesis (Fig 4) as to how the enol ether arises. The substituted dihydrobenzofuran core likely arise through radical coupling of the single-electron oxidation products of 5 and 9 followed by nucleophilic attack of the C-3 hydroxyl group in 5 on C-7 of 9 to generate 12 (Dewick, 2002) (Fig 4A). The carbons of the 2-(3,5-dihydroxyphenyl)ethenyl ether found in 4 originate via an acetate-derived pathway, while the 2-(3,4-dihydroxyphenyl)ethenyl ether found 3 in arise via a more traditional shikimic acid biosynthesis. On causal inspection it appears these moieties then couple with the corresponding aldehyde acting a nucleophile through its enol form, a rather unlikely proposal. To avoid this issue, Brown and coworkers (Grayer et al., 2003) originally hypothesized an addition, esterification, and elimination sequence involving a transient biogenetic equivalent of the aldehyde 13 in the biosynthesis of 6 and 7 (Fig 4B). This intermediate (13) reacts with a thiol to generate 14 after which the alcohol is esterified (15) and the thiol eliminated (16). An alternatively proposal is shown in Fig 4C, which involves simple esterification of the corresponding α-hydroxyacid (17), found in 3 and 4 already, followed by oxidative decarboxylation (Boland and Mertes, 1985; Neumann and Boland, 1990; Dewick, 2002). Stereospecific loss of the pro-R or pro-S methylene proton during this process would result in either E- or Z-olefin.
Fig. 4.
Proposed Biosynthesis of A) benzodihydrofuran core 12 B) enol ether in 6 and 7 (Grayer et al., 2003) C) enol ether in 3 and 4 on the basis of oxidative decarboxylation.
2.2 Biological activity evaluation
Sebestenoids A-D were tested for their ability to inhibit β-secretase. In the Amyloid Cascade Hypothesis (Hardy, 2006) of Alzheimer's disease progression, cleavage of amyloid precursor protein (APP) by BACE1 begins a cascade leading to the formation of soluble polypeptide oligomers that trigger neurodegeneration (Shankar et al., 2008). Compounds 3 and 4 were the most potent with IC50 values of 20 and 22 μM, respectively. In comparison, the smaller compounds 1 and 2 were less active at 32 and 116 μM, respectively.
Phenylpropanoids have been reported to display a wide range of activities, raising questions regarding the specificity of 3 and 4. Recently, Shoichet et al have demonstrated that many compounds that frequently hit in assays are promiscuous inhibitors which aggregate with enzymes in a nonstoichiometric fashion (Feng and Shoichet, 2006), which disrupts protein folding (Coan et al., 2009). To determine if the observed effects were due to aggregation, our BACE1 assay was repeated with the addition of detergent to disrupt these non-specific interactions. Unfortunately, our original assay is unable to tolerate the suggested conditions (0.01 or 0.1% Triton). Presumably, this additive is also disrupting the crucial protein aggregation step in our complementation assay (Eglen, 2002). The activities of 3 and 4 were therefore investigated using a surrogate system. The compounds were assayed against the serine protease chymotrypsin in a standard chemiluminescent assay (Gunasekera et al., 2010) with and without the addition of detergent. The IC50 values of both compounds were strongly affected by this additive. For 3, the IC50 value was 33 μM without detergent but in the presence of 0.01% Triton (v/v) the potency dropped to 134 μM. The effect with compound 4 was similar, as this compoundinhibited chymotrypsin with an IC50 value of 27 μM, which increased to 65 μM in the presence of detergent. This behavior is similar to that observed for Congo Red in this same assay, which is a known aggregator, suggesting that these compounds also interacting with both enzymes in a similar non-specific fashion.
3. Conclusion
From a Hawaiian sample of C. sebestena four new metabolites have been isolated and their structures defined. Compounds 2 and 4 are relatively rare examples of 2-(3,5-dihydroxyphenyl)ethenyl ethers. In our hands these units were unstable and degraded to complex mixtures rapidly. This finding may partially explain why there are so few reports of compounds containing this moiety. Biological evaluation against serine and aspartic proteases showed that these compounds inhibited both enzymes. While this activity was dose-dependent, it was also strongly influence by the addition of detergents suggesting a non-specific inhibition. The operational simplicity of this counter screen suggests it should be incorporated early in any screening campaign involving plant natural products.
4. Experimental
4.1 General experimental procedures
Optical rotations were measured on a polarimeter at the sodium line (589 nm). UV spectra were obtained on a spectrophotometer and IR spectra were measured as a thin film on a CaF2 disk. NMR spectra were acquired on a 500 MHz spectrometer operating at 500 (1H) or 125 (13C) MHz using the residual solvent signals as an internal reference (CD3OD δH 3.30 ppm, δC 49.0 ppm). High-resolution mass spectroscopic data were obtained on a LC-MSTOF with ES ionization in the positive mode. Gradient separations were performed on a system consisting of solvent delivery modules, a photodiode diode array detector, an evaporating light scattering detector, and a system controller. TLC analyses was performed on Si60F254 plates and visualized under UV or by heating after spraying with a 1% anisaldehyde solution in acetic acid/H2SO4 (50:1). Rosmarinic acid was purchased from Sigma-Aldrich.
4.2. Plant Material
The fruits of Cordia sebestena were collected in Honolulu, Hawaii, in August 2009 adjacent to the University of Hawaii at Manoa campus. A voucher specimen (No. 0901) is maintained in the Department of Chemistry at University of Hawaii at Manoa.
4.3. Extraction and Isolation
The fruits (2.5 kg) were extracted thrice with ethyl acetate. The EtOAc extract (10 g) was separated over silica gel (300-400 mesh) with a step-gradient of increasing amounts of ethyl acetate in hexane (1:10, 1:7, 1:5, 1:3, 1:1, 1:0). The gummy fraction (Fraction 15) containing 1 and 2 eluted with 1:1 hexanes:EtOAc, while compounds 3 and 4 eluted with 100% EtOAc (Fraction 17). Fraction 15 (200 mg) was separated by RP-HPLC [Luna C8, 250 × 10 mm, a linear gradient from 5-100% MeCN in water over 40 min, flow rate 3 mL/min, PDA and ELSD detection] to afford sebestenoid A (1) (tR 26.0 min, 2.0 mg) and sebestenoid B (2) (tR 25.0 min, 1.0 mg). Sebestenoid C (3) (tR 24.0 min, 2.0 mg) and sebestenoid D (4) (tR 23.5 min, 1.0 mg) were isolated from Fraction 17 (150 mg) by RP-HPLC [Luna C8, 250 × 10 mm, a linear gradient from 5-100% MeOH in water over 40 min, flow rate 3 mL/min, PDA and ELSD detection].
4.4. Characterization
4.4.1. Sebestenoid A (1)
Amorphous light yellow powder; UV (MeOH) λmax (log ε) 243 (4.1), 252 (4.1), 301 (4.1), 334 (4.2), 343 (4.2) nm; IR (CaF2) νmax 3403, 1702, 1603, 1504, 1260, 1123 cm-1; For 1H (500 MHz, MeOH-d4) and 13C (125 MHz, MeOH-d4) NMR spectroscopic data, see Tables 1 and 2; HRESI-TOFMS obsd m/z: 507.1288 [M + H]+, calcd for C27H23O10+, 507.1291.
Table 2.
13C NMR Spectroscopic Data for Compounds 1-4 (125 MHz, δ ppm)
| Position | 1a | 2a | 3a | 4a |
|---|---|---|---|---|
| 1 | 127.7 | 127.3 | 127.8 | 127.5 |
| 2 | 117.3 | 116.6 | 117.4 | 116.5 |
| 3 | 146.1 | 146.3 | 146.0 | 146.2 |
| 4 | 148.5 | 113.6 | 145.7 | 113.6 |
| 5 | 116.1 | 146.5 | 116.5 | 146.3 |
| 6 | 122.7 | 119.6 | 122.8 | 119.6 |
| 7 | 113.3 | 116.8 | 113.2 | 116.8 |
| 8 | 132.8 | 135.4 | 132.9 | 135.5 |
| 1' | 130.8 | 130.8 | 124.5 | 124.4 |
| 2' | 116.8 | 116.8 | 126.5 | 126.3 |
| 3' | 145.5 | 146.6 | 149.1 | 149.2 |
| 4' | 148.4 | 148.8 | 145.9 | 145.7 |
| 5' | 115.5 | 115.5 | 118.6 | 118.6 |
| 6' | 122.3 | 122.3 | 122.2 | 122.3 |
| 7' | 148.0 | 147.6 | 144.9 | 144.5 |
| 8' | 115.8 | 115.7 | 115.7 | 115.7 |
| 9' | 165.4 | 165.9 | 165.5 | 165.9 |
| 1" | 125.4 | 125.4 | 133.5 | 133.5 |
| 2" | 118.2 | 118.1 | 113.3 | 113.3 |
| 3" | 146.1 | 148.0 | 146.1 | 146.5 |
| 4" | 149.2 | 149.2 | 146.8 | 146.6 |
| 5" | 116.4 | 116.4 | 116.3 | 116.4 |
| 6" | 125.2 | 125.2 | 118.3 | 118.3 |
| 7" | 129.8 | 129.8 | 88.1 | 88.2 |
| 8" | 138.3 | 138.2 | 57.4 | 57.6 |
| 9" | 165.6 | 165.5 | 172.3 | 172.3 |
| 1"' | 128.5 | 128.5 | ||
| 2"' | 117.2 | 117.3 | ||
| 3"' | 146.6 | 146.8 | ||
| 4"' | 145.2 | 145.3 | ||
| 5"' | 116.5 | 116.5 | ||
| 6"' | 121.8 | 121.8 | ||
| 7"' | 37.4 | 37.5 | ||
| 8"' | 75.7 | 75.7 | ||
| 9"' | 171.2 | 171.2 | ||
| OCH3 | 52.8 | 52.8 | 52.8 | 52.9 |
Data measured in MeOH-d4
b Compounds numbered as in Murata et al (2009).
4.4.2. Sebestenoid B (2)
Light yellow powder; UV (MeOH) λmax (log ε) 333 (4.0), 292 (3.9) nm; IR (CaF2) νmax 3385, 1709, 1598, 1501,1441, 1259, 1122 cm-1; For 1H (500 MHz, MeOH-d4) and 13C (125 MHz, MeOH-d4) NMR spectroscopic data, see Tables 1 and 2; HRESI-TOFMS obsd m/z: 507.1290 [M + H]+, calcd for C27H23O10+, 507.1291.
4.4.3. Sebestenoid C (3)
Light yellow powder; [α]D22 +32 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 335 (4.3), 290 (4.2), 257 (4.3) nm; IR (CaF2) νmax 3401, 1728, 1606, 1504, 1440, 1280, 1155cm-1; For 1H (500 MHz, MeOH-d4) and 13C (125 MHz, MeOH-d4) NMR spectroscopic data, see Tables 1 and 2; HRESI-TOFMS obsd m/z: 709.1532 [M + Na]+, calcd for C36H30O14Na+, 709.1533.
4.4.4. Sebestenoid D (4)
Light yellow powder; [α]D22 +77 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 223 (4.7), 253 (4.4), 292 (4.4), 337 (4.4) nm; IR (CaF2) νmax 3398, 1730, 1608, 1508, 1260, 1176 cm-1; For 1H (500 MHz, MeOH-d4) and 13C (125 MHz, MeOH-d4) NMR spectroscopic data, see Tables 1 and 2; HRESI-TOFMS obsd m/z: 709.1526 [M + Na]+, calcd for C36H30O14Na+, 709.1533.
4.4.5. Alkaline Hydrolysis of Rosmarinic Acid (8)
Rosmarinic acid (100 mg) was dissolved in 10% NaOH (5 mL) and the mixture stirred for 1 h at room temperature under N2. The reaction mixture was passed through a DOWEX 50W-X2 column (3 × 6 cm) and eluted with H2O (200 mL). The eluate was concentrated under reduced pressure to give a brown residue (50 mg). The residue was then purified by RP-HPLC [Luna C8, 250 × 10 mm, a linear gradient from 5-10% MeCN in water (0.1% formic acid) over 40 min, flow rate 3 mL/min, PDA and ELSD detection] to give (2R)-3-(3,4-dihydroxyphenyl)-2-hydroxypropanoic acid (4.2 mg) as a colorless, amorphous solid: [α]D22 +13 (c 0.2, MeOH); 1H NMR (MeOH-d4) δH 2.74 (1H, dd, J = 13.8, 7.8 Hz), 2.94 (1H, dd, J = 13.8, 4.5 Hz), 4.24 (1H, dd, J =7.8, 4.5 Hz), 6.56 (1H, dd, J = 8.1, 2.1 Hz), 6.66 (1H, d, J = 8.1 Hz), 6.71 (1H, d, J = 2.1 Hz).
4.4.6. Alkaline Hydrolysis of a Mixture of Sebestenoid C (3) & D (4)
A mixture of sebestenoids C (3) and D (4) (6.0 mg) was dissolved in 10% NaOH (0.5 mL) and the mixture stirred for 2 h at room temperature under N2. The reaction mixture was passed through a DOWEX 50W-X2 column (2 × 5 cm) and eluted with H2O (100 mL). The eluate was concentrated under reduced pressure to give a brown residue (5.0 mg). The residue was purified by RP-HPLC [Luna C8, 250 × 10 mm, a linear gradient from 5-50% MeCN in water (0.1% formic acid) over 40 min, flow rate 3 mL/min, PDA and ELSD detection] to give (2R)-3-(3-methoxy-4-hydroxyphenyl)-2-hydroxypropanoic acid (10, 1.1 mg) as a colorless, amorphous solid, [α]D22 +13 (c 0.2, MeOH); 1H NMR (MeOH-d4) δH 2.71 (1H, dd, J = 13.8, 7.8 Hz), 2.93 (1H, dd, J = 13.8, 4.2 Hz), 4.20 (1H, dd, J =7.8, 4.2 Hz), 6.57 (1H, dd, J = 8.1, 1.6 Hz), 6.65 (1H, d, J = 8.1 Hz), 6.68 (1H, d, J = 1.6 Hz), and (2S,3S)-4-((E)-2-carboxyvinyl)-2-(3,4-dihydroxyphenyl)-7-hydroxy-2,3-dihydrobenzofuran-3-carboxylic acid (11, 1.8 mg), as a colorless, amorphous solid, [α]D22 +137 (c 1.1, MeOH), +168 (c 0.2, MeOH); 1H NMR (MeOH-d4) δH 4.27 (1H, d, J = 5.1 Hz), 5.86 (1H, d, J = 5.1 Hz), 6.26 (1H, d, J = 15.9 Hz), 6.72 (1H, d, J = 8.0 Hz), 6.72 (1H, d, J = 8.0 Hz), 6.79 (1H, d, J = 2.1 Hz), 6.77 (1H, dd, J = 8.0, 2.1 Hz), 7.15 (1H, d, J = 8.0 Hz), 7.78 (1H, d, J = 15.9 Hz); HRESI-TOFMS obsd m/z 381.0586 [M + Na]+, calcd for C18H14O8Na+, 381.0586.
4.4.7 Protease Assay
The proteolytic cleavage of amyloid precursor protein was assayed as described (Naqvi, 2004). Test compounds were dissolved in DMSO at the desired concentration and incubated in triplicate with the enzyme for 16 h in 96 well plates. A DMSO control (1.5 μL) and an inhibitor standard were also tested in triplicate. The chemiluminescence signal was read using a Fluostar Optima spectrophotometer. Data were analyzed using GraphPad Prism. BACE1 activity was calculated as a percent of the positive control using a nonlinear regression analysis function that corresponded to a best one-fit model.
Supplementary Material
Acknowledgment
This work was funded by grants from the Victoria S. and Bradley L. Geist Foundation (20070461), the Alzheimer's Association NIRG-08-90880, Alzheimer's Drug Discovery Foundation (281204), the National Institute of Aging (R20072671). Funds for the upgrades of the NMR instrumentation were provided by the CRIF program of the National Science Foundation (CH E9974921) and the Elsa Pardee Foundation. The purchase of the Agilent LC-MS was funded by grant W911NF-04-1-0344 from the Department of Defense. We thank T. Hemscheidt for helpful discussions regarding the biosynthesis and B. Rubio for the chymotrypsin data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- Abbott IA. La'Au Hawaii: Traditional Hawaiian Uses of Plants. 1st ed. Bishop Museum Press; 1992. [Google Scholar]
- Arihara S, Rüedi P, Eugster CH. Dopaldehyd: erstmalige Isolierung aus einer Pflanze in Form seines Enol--Kaffeesäureesters. Helv. Chim. Acta. 1975;58:447–453. [Google Scholar]
- Boland W, Mertes K. Biosynthesis of algal pheromones. Eur. J. Biochem. 1985;147:83–91. doi: 10.1111/j.1432-1033.1985.tb08722.x. [DOI] [PubMed] [Google Scholar]
- Burney DA, James HF, Burney LP, Olson SL, Kikuchi W, Wagner WL, Burney M, McCloskey D, Kikuchi D, Grady FV, Gage R, Nishek R. Fossil Evidence For A Diverse Biota From Kaua`i and its Transformation Since Human Arrival. Ecol. Monogr. 2001;71:615–641. [Google Scholar]
- Chun MN, Hawaii . Native Hawaiian Medicines, A new revised and enlarged translation. First People's Productions; Honolulu, Hawaii: 1994. [Google Scholar]
- Chun MN, Kapunihana JK. Native Hawaiian Medicine. First People's Productions; Honolulu: 1998. [Google Scholar]
- Coan KED, Maltby DA, Burlingame AL, Shoichet BK. Promiscuous Aggregate-Based Inhibitors Promote Enzyme Unfolding. J. Med. Chem. 2009;52:2067–2075. doi: 10.1021/jm801605r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. 2nd ed. Wiley; 2002. [Google Scholar]
- Eglen RM. Enzyme fragment complementation: a flexible high throughput screening assay technology. Assay Drug Dev. Techn. 2002;1:97–104. doi: 10.1089/154065802761001356. [DOI] [PubMed] [Google Scholar]
- Euranto E. Mechanisms and Catalysis in Vinyl Ester Hydrolysis. Pre & Appl. Chem. 1977;49:1009–1020. [Google Scholar]
- Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayer RJ, Eckert MR, Veitch NC, Kite GC, Marin PD, Kokubun T, Simmonds MS, Paton AJ. The chemotaxonomic significance of two bioactive caffeic acid esters, nepetoidins A and B, in the Lamiaceae. Phytochemistry. 2003;64:519–528. doi: 10.1016/s0031-9422(03)00192-4. [DOI] [PubMed] [Google Scholar]
- Gunasekera SP, Miller MW, Kwan JC, Luesch H, Paul VJ. Molassamide, a depsipeptide serine protease inhibitor from the marine cyanobacterium Dichothrix utahensis. J. Nat. Prod. 2010;73:459–462. doi: 10.1021/np900603f. [DOI] [PubMed] [Google Scholar]
- Gutmanis J. Kahuna La□ au Lapa□ au: The Practice of Hawaiian Herbal Medicine. Island Heritage; Aiea, Hawaii: 1976. [Google Scholar]
- Hardy J. Has the Amyloid Cascade Hypothesis for Alzheimers Disease been Proved? Curr. Alzheimer Res. 2006;3:71–73. doi: 10.2174/156720506775697098. [DOI] [PubMed] [Google Scholar]
- Kaaiakamanu DM, Chun MN. Native Hawaiian Medicine. First People's Productions; Honolulu, H.I.: 2003. [Google Scholar]
- Kaitin KI. Deconstructing the Drug Development Process: The New Face of Innovation. Clin. Pharmacol. Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss BH. Plants in Hawaiian Culture. 1st ed. University of Hawaii Press; Honolulu: 1993. [Google Scholar]
- Murata T, Sasaki K, Sato K, Yoshizaki F, Yamada H, Mutoh H, Umehara K, Miyase T, Warashina T, Aoshima H, Tabata H, Matsubara K. Matrix Metalloproteinase-2 Inhibitors from Clinopodium chinense var. parviflorum. J. Nat. Prod. 2009;72:1379–1384. doi: 10.1021/np800781t. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Nishi M, Inada A, Obata H, Tanabe N, Abe S, Wakashiro M. Two new potent inhibitors of xanthine oxidase from leaves of Perilla frutescens Britton var. acuta Kudo. Chem. Pharm. Bull. 1990;38:1772–1774. doi: 10.1248/cpb.38.1772. [DOI] [PubMed] [Google Scholar]
- Naqvi T. Galactosidase Enzyme Fragment Complementation as a High-Throughput Screening Protease Technology. J. Biomol. Screen. 2004;9:398–408. doi: 10.1177/1087057104264040. [DOI] [PubMed] [Google Scholar]
- Neumann C, Boland W. Stereochemical studies on algal pheromone biosynthesis. Eur. J. Biochem. 1990;191:453–459. doi: 10.1111/j.1432-1033.1990.tb19143.x. [DOI] [PubMed] [Google Scholar]
- Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Kido T, Tanaka N, Murakami T, Saiki Y, Chen C-M. Chem. Pharm. Bull. 1992;40:2099–2101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.