Abstract
The RECODE database is a compilation of ‘programmed’ translational recoding events taken from the scientific literature and personal communications. The database deals with programmed ribosomal frameshifting, codon redefinition and translational bypass occurring in a variety of organisms. The entries for each event include the sequences of the corresponding genes, their encoded proteins for both the normal and alternate decoding, the types of the recoding events involved, trans-factors and cis-elements that influence recoding. The database is freely available at http://recode.genetics.utah.edu/.
INTRODUCTION
Recoding is the reprogramming of mRNA translation by localized alterations in the standard translational rules. Recoding is utilized in the expression of a minority of genes in probably all organisms, though the extent of occurrence within organisms is unknown. In known cases the product of recoding is functionally important, but there may exist cases where the importance of recoding does not lie in the encoded protein. Three classes of recoding are known.
(i) Frameshifting at a particular site can yield two protein products from one coding sequence or one protein product from two overlapping open reading frames (ORFs). In some cases a set ratio of two products is important and in other cases frameshifting has a regulatory purpose. In the latter the level of frameshifting is influenced by the concentration of proteins or other factors present during translation. The known cases of frameshifting where the product is utilized involve shifts of one base, either +1 or –1 (but shifts of two bases have been demonstrated in artificial systems and changes of reading frame may also occur with bypassing).
(ii) Bypassing (hopping) occurs when a block of nucleotides within a coding sequence is not translated. Translation is temporarily suspended, ribosomes traverse the coding gap and protein synthesis resumes to yield a single protein. This allows the coupling of two ORFs separated on an mRNA by a coding gap, and is frame-independent.
(iii) Codon redefinition involves site-specific alteration of codon meaning. All the included cases involve redefinition of a stop codon to specify an amino acid, often glutamine, tryptophan or selenocysteine. (The altered meaning of certain codons when they function as an initiation codon is not included in this compendium.)
An example of each type is illustrated in Figure 1 and independent cases of recoding are summarized in Table 1. Recoding events occur in competition with standard readout of the transcript, and are site specific. The efficiency of recoding at the site is usually influenced by stimulatory signals present on the mRNA (cis-elements) and in some cases by protein products or other cellular components (trans-elements). Detailed information about recoding and its mechanisms can be found elsewhere (1–3)
Figure 1.
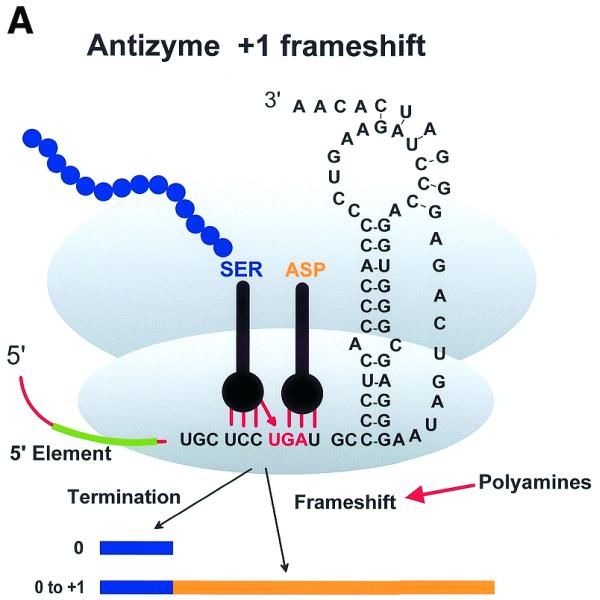
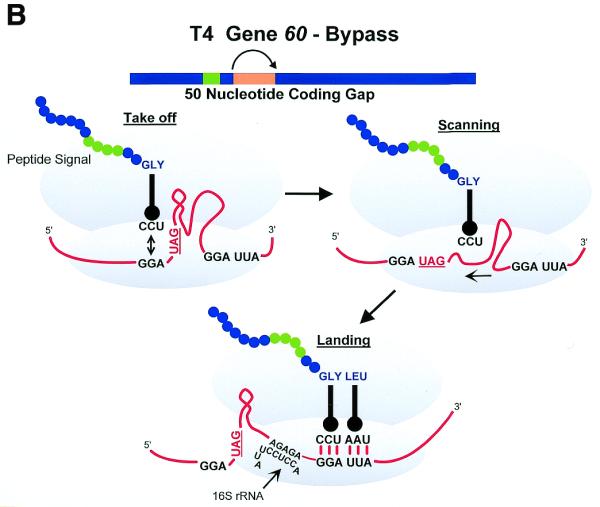
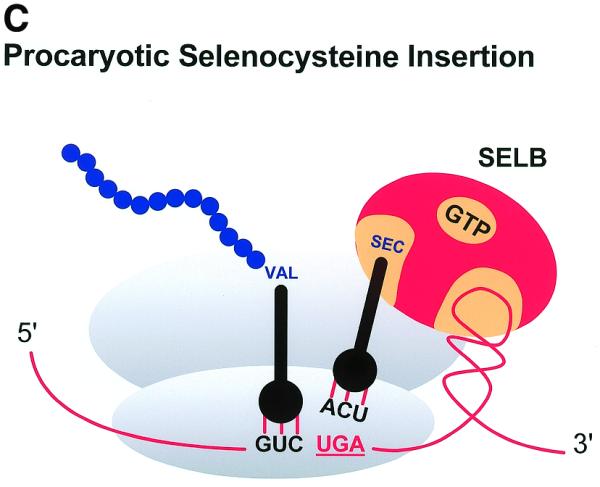
Three examples of recoding events. (A) Antizyme frameshifting. The +1 shift at the last codon (UCC) before the termination codon of ORF1 of human antizyme 1 is stimulated by polyamines and by a 5′ mRNA element and a 3′ pseudoknot. (B) Gene 60 bypassing. Fifty nucleotides between codons 47 and 48 of phage T4 gene 60 coding sequence are bypassed by half the ribosomes in response to matched takeoff and landing site codons, a stop codon directly after the take-off site in a stem–loop structure and a nascent peptide signal that acts within the ribosome. (C) Procaryotic selenocysteine insertion—redefinition. UGA codons in procaryotes that specify selenocysteine are directly followed by a stem structure whose apical loop is bound by a selenocysteine tRNA specific elongation factor SELB, resulting in a tethered aminoacylated tRNA poised for the oncoming ribosome. These figures are adapted from Atkins et al. (8).
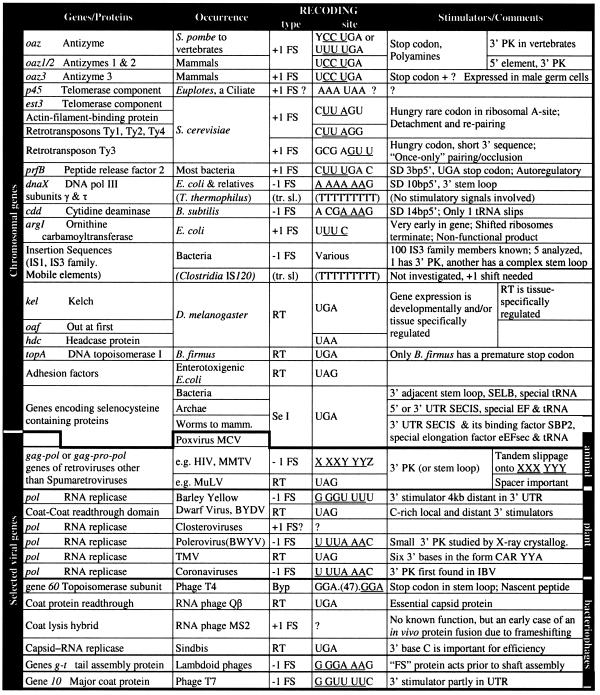
Table 1. An overview of gene types that utilize recoding for their expression.
References are in reviews (1–3) and in the database.
FS, frameshifting; RT, readthrough (codon redefinition); Se I, selenocysteine insertion (a special case of redefinition); Byp, translational bypasing (the matched takeoff and landing site codons, GGA, in T4 gene 60 bypassing are separated by 47 nt, indicated in parenthesis); PK, RNA pseudoknot; EF, elongation factor. SD 3bp5′ indicates that the distance between Shine–Dalgarno sequence, with which translating ribosomes interact to influence recoding, is 3 base pairs 5′ of the frameshift site. Codons in the initial frame are separated by spaces and the codons at which the new frame is set are underlined. Two cases of transcription slippage (tr. sl.) are also included as they yield the same end result as recoding. In these cases the DNA sequence on which the RNA polymerase slips is given.
Currently a huge increase of genomic sequence information is being obtained from many different organisms. Genome annotations provide information about many observed and putative products, but those resulting from recoding are often overlooked. It is clear that a number of recoding products play critical cellular roles. Thus it is important to have a comprehensive, accurate and consistent resource that provides recoding event information.
DESCRIPTION OF THE DATABASE
The data are stored using an SQL based relational database (OpenBase, Francetown, NH) and mapped to the Web using WebObjects middleware (Apple Enterprise, Cupertino, CA). The data are organized and stored relationally to allow flexibility in annotation and ease of future enhancements such as complex queries. Currently, data are presented to the user with relational information joined into a unified view of individual recoding events. In late 2000 the database consisted of 227 recoding events. A forms-based search mechanism is provided to allow specification of recoding category, organism, gene name, product(s) plus its function and cis- and trans-elements involved. Searches in all non-sequence fields are also possible. Search fields left blank are taken as wildcards, and all string-based fields accept the wildcards ‘*’ (match 0 or more characters) and ‘?’ (match any single character). Entries have the following fields.
(i) Gene, common gene name.
(ii) Type, type of recoding event (see Introduction).
(iii) Organism name, official name of the organism and other names used. Other names are represented according to information provided by the NCBI Taxonomy Database (4).
(iv) Cis-elements, characterized elements of primary, secondary or tertiary mRNA structure that are known to influence recoding.
(v) Trans-elements, cellular factors that influence recoding, e.g. proteins, small ligands etc.
(vi) Function of recoding, recoding can play various roles in cells. It is utilized for the regulation of gene expression level such as in the synthesis of bacterial release factors 2 (RF2) (5) or in eukaryotic antizymes (OAZ) (6). Recoding is also utilized for production of the proper ratio between two proteins; an example is the expression of Escherichia coli dnaX where two subunits of DNA polymerase III (γ and τ) are synthesized in a 1:1 ratio (7).
(vii) Product/function, information about a gene product(s) and its/their function(s).
(viii) Translation without recoding, sequence of the protein or polypeptide synthesized by standard translation.
(ix) Translation with recoding, sequence of the protein whose synthesis results from recoding.
(x) References, primary research papers that describe particular recoding events are cited with the corresponding hyperlink to their MEDLINE abstract.
(xi) Comments/notes, any important information about the entry that is not described in others fields.
(xii) Evidence, provides information on whether the (presumed) event has been demonstrated experimentally.
(xiii) Gene/mRNA, nucleotide sequence of the mRNA.
Because many mRNAs are very large, only the gene sequence of interest is given. For each gene the translation initiation codon as well as the mRNA elements thought to be important for recoding are marked. The following designations are used in the database: bold, any regions of mRNA which are important for recoding; underlined, recoding site (frameshift site for frameshift events, take-off and landing-site codons for translational bypass, redefined codon for codon redefinition); blue, start codon; red, stop codons important for recoding; brown, Shine–Dalgarno sequence; green and violet, double-stranded RNAs that play a role in frameshifting, violet is for stem 2 of pseudoknots and kissing loops; sequences in italics, those whose importance for recoding is at the level of their peptide product. The logo of the database (Fig. 2) can be used as a key for these designations
Figure 2.

Logo of the database.
AVAILABILITY AND SUMISSIONS
RecodeWeb is freely available at http://recode.geneti cs.utah.edu/. The authors welcome submission of new examples or additional information about already entered recoding events. Submission should be via electronic form at http://re code.genetics.utah.edu/submission/.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Ivaylo Ivanov and Stefan Aigner for providing data before publication, and the following sources of support for this work: National Cancer Center fellowship to P.V.B.; Thomas Dee fellowship to O.L.G.; Centre National de la Recherche Scientifique, the Université Paul Sabatier and a grant from the Programme de Recherche Fondamentale en Microbiologie et maladies infectieuses et parasitaires to O.F.; grant (DEFG03-99ER62732) from Department of Energy to R.F.G; NIH grant GM 48152 to J.F.A. and NHGRI Genome Scholar award (K22 HG000401) to M.C.G.
References
- 1.Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- 2.Farabaugh P.J. (1996) Programmed translational frameshifting. Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- 3.Miller W.A., Brown,C.M. and Wang,S. (1997) New punctuation for the genetic code: Luteovirus gene expression. Semin. Virol., 8, 3–13. [Google Scholar]
- 4.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. Updated article in this issue: Nucleic Acids Res. (2001), 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craigen W.J. and Caskey,C.T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- 6.Matsufuji S., Matsufuji,T., Miyazaki,Y., Murakami,Y., Atkins,J.F., Gesteland,R.F. and Hayashi,S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell, 80, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen B., Gesteland,R.F. and Atkins,J.F. (1997) Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol., 271, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.