Abstract
The PlantsP database is a curated database that combines information derived from sequences with experimental functional genomics information. PlantsP focuses on plant protein kinases and protein phosphatases. The database will specifically provide a resource for information on a collection of T-DNA insertion mutants (knockouts) in each protein kinase and phosphatase in Arabidopsis thaliana. PlantsP also provides a curated view of each protein that includes a comprehensive annotation of functionally related sequence motifs, sequence family definitions, alignments and phylogenetic trees, and descriptive information drawn directly from the literature. PlantsP is available at http://PlantsP.sdsc.edu.
INTRODUCTION
As the first complete genomic sequence of a plant, Arabidopsis thaliana, approaches completion, scientific focus is shifting from the genomic sequence to functional genomics, the understanding of how genes encoded in the genome interact to produce the complex characteristics and phenotypes of the intact organism. The genomic sequence provides a framework for organizing functional genomic information not only for Arabidopsis, but for other plants as well. The PlantsP database provides such a framework for proteins involved in phosphorylation, i.e. protein kinases, protein phosphatases and their substrates. In addition, PlantsP is a community resource designed to collect annotations and descriptive information from participants in the project and from the scientific community as a whole.
Although the Arabidopsis sequence provides a framework for the data, PlantsP comprises information from all plant species (i.e. kingdom viridiplantae). When the Arabidopsis sequence is complete, it will be possible to examine the relationship of plant protein kinases and protein phosphatases to their probable orthologs in Arabidopsis.
DATABASE CONTENT
The genomic sequence of Arabidopsis provides a basis for organizing the functional genomic data that will be produced over the next few years. Hence, the initial focus of PlantsP has been on identifying and annotating a comprehensive and non-redundant set of protein kinase and protein phosphatase genomic sequences, cDNA sequences and protein sequences. This work is still in progress (Table 1) as the Arabidopsis sequence is not yet complete. While it would seem straightforward to assemble a unique set of genomic sequences, cDNA transcript sequences and the encoded protein sequences, it is in fact quite difficult because of duplications and incomplete referencing of related sequences in the public databases. A significant amount of curatorial effort is required to correctly identify related sequences and create a single merged entry. The alternative to this merging process, to base the PlantsP archive on only a subset of the sequences in the public database is unpalatable because significant amounts of source annotation would be lost in such an approach.
Table 1. Content of selected tables in PlantsP database.
| Database table |
Purpose |
Number of entries |
| uid | Records unique identifier for sequences, structures, features, annotations and cross-references | 8371 |
| sequence_protein | Protein sequence | 2546 |
| sequence_rna | Nucleotide sequence of coding region | 363 |
| sequence_genomic | Nucleotide sequence of genomic region ± 1000 bases around coding sequence | 368 |
| gene-structure | Exon/intron/UTR structure of genes (note there may be multiple gene structures) | 2423 |
| gene_region | Genes in the region of each genomic sequence | 366 |
| knockouts | T-DNA insertional knockouts isolated in this project | 33 |
| annotation_basic | Basic annotation | 3504 |
| citation | Literature citations for sequences, features, feature methods, annotations, etc. | 4102 |
| feature | Pattern descriptions used to identify sequence features, e.g. domains, motifs, etc. | 214 |
| feature_method | Computational methods used to identify sequence features | 3 |
| feature_instance | Occurrences of sequence features in particular sequences | 8875 |
| xref | Cross-references to external databases such as GenBank, EMBL, PDB, SWISS-PROT, PIR, etc. | 5108 |
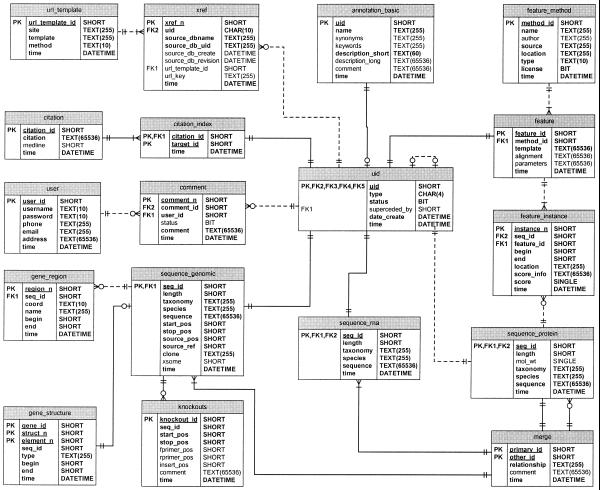
The PlantsP database schema (Fig. 1) is designed to hold detailed annotation supporting the reasoning used in assigning sequence features and putative functions. For sequence features, this information is found in the feature_method table which describes the computational method used to make a feature assignment (including a literature reference), and the feature table which contains the actual pattern description (e.g. signature, profile or HMM) used to identify the feature and the threshold values used. Occurrences of features in the feature_instance table also include the score so that the user can evaluate the significance of the identification of the feature.
Figure 1.
PlantsP database design.
The PlantsP database is a component of a functional genomics project, ‘Functional genomics of plant phosphoproteins’. The experimental side of this project involves screening for T-DNA insertional knockouts for the Arabidopsis protein kinases and protein phosphatases identified in the PlantsP database. As insertional mutations are identified, their locations are included in the knockouts table; seed stocks from each insertion line are available from the Arabidopsis Biological Resource Center (see below).
The knockout screening process involves several rounds of screening of DNA from pooled knockout lines, which is performed using the services of the Arabidopsis Knockout Facility (1). After screening, seeds must be grown in bulk for deposition with the ABRC (Arabidopsis Biological Resource Center, 309 B&Z Building, 1735 Neil Avenue, Columbus, OH 43210, USA; http://aims.cps.msu.edu/aims/). The experimental portion of the project has just reached the point of identifying the first knockout lines and substantial additions will be made to accommodate new fields describing the phenotypes of mutants. The knockout information will be used as a prototype for other kinds of functional genomic information as it becomes available (such as expression profiling, substrates and protein–protein interaction results).
The PlantsP schema includes tables for recording both user annotations (comment table) as well as for tracking the submitter of annotations (user table). These annotations will appear in the database with a citation identifying the contributor. The user table will also be used in the future to implement server-push functionality. Server-push functions allow the database to notify the user when specific events occur. Examples of server-push function include notifying the user when a knockout has been found in a specific gene, when a new gene product containing a set of specified motifs is added to the database or when the annotation has been updated on a specified set of genes.
In addition to the information available in the relational database, PlantsP also includes curated information related to the classification of protein kinases and protein phosphatases in plants, gene-based phylogenetic trees showing the interrelationships of kinases and phosphatases, and a selective index of recently published papers related to plant phosphorylation.
ACCESS
PlantsP is available on the web at http://PlantsP.sdsc.edu. A simple interface allowing searching based on keywords, and limited by species and molecular weight is currently available. Queries specifying logical combinations of known sequence and structural motifs can be made, as well as searches using a known sequence as a query using the BLAST program (2). These approaches give a simple but flexible access to the data and will be expanded in the future.
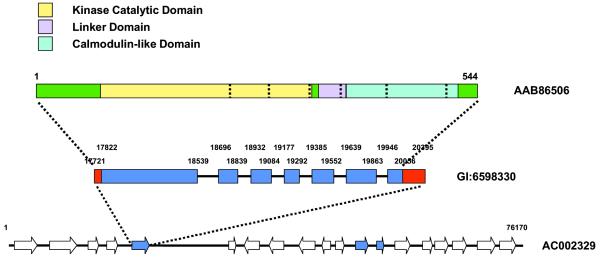
The results of queries to the database are currently displayed as hyperlinked text. A graphical display is currently being implemented, and should be available by early 2001 (Fig. 2).
Figure 2.
Graphical display of sequence features. AAB86506 represents a protein sequence with functional regions shown in color and intron/exon boundaries shown as dotted lines. GI:6598330 represents a primary transcript with untranslated regions (red), exons (blue) and introns (black). AC002329 show genes in the region of the coding sequence with unknown genes (white), known genes (hatched) and other kinase or phosphatase genes (blue).
FUTURE DEVELOPMENT PLANS
• Complete incorporation of all Arabidopsis protein kinases and protein phosphatases based on the completed genomic sequence.
• Complete sequence based classification of kinases.
• Complete annotation of functional and conserved sequence domains.
• Implementation of graphical display for both single and multiple sequence query responses.
• Complete linkage of all plant protein kinases and protein phosphatases to Arabidopsis orthologs.
• Extension to other kinds of functional genomic information, especially to protein–protein interactions, substrates and expression profiling.
Acknowledgments
ACKNOWLEDGEMENT
The authors are supported by funding from the National Science Foundation, DBI-9975808.
References
- 1.Krysan P.J., Young,J.K. and Sussman,M.R. (1999). T-DNA as an Insertional Mutagen in Arabidopsis. Plant Cell, 2, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]