Abstract
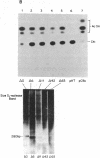
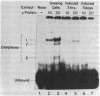
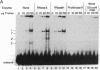
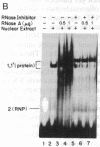
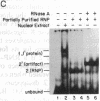
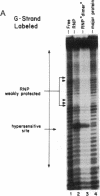
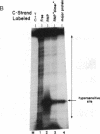
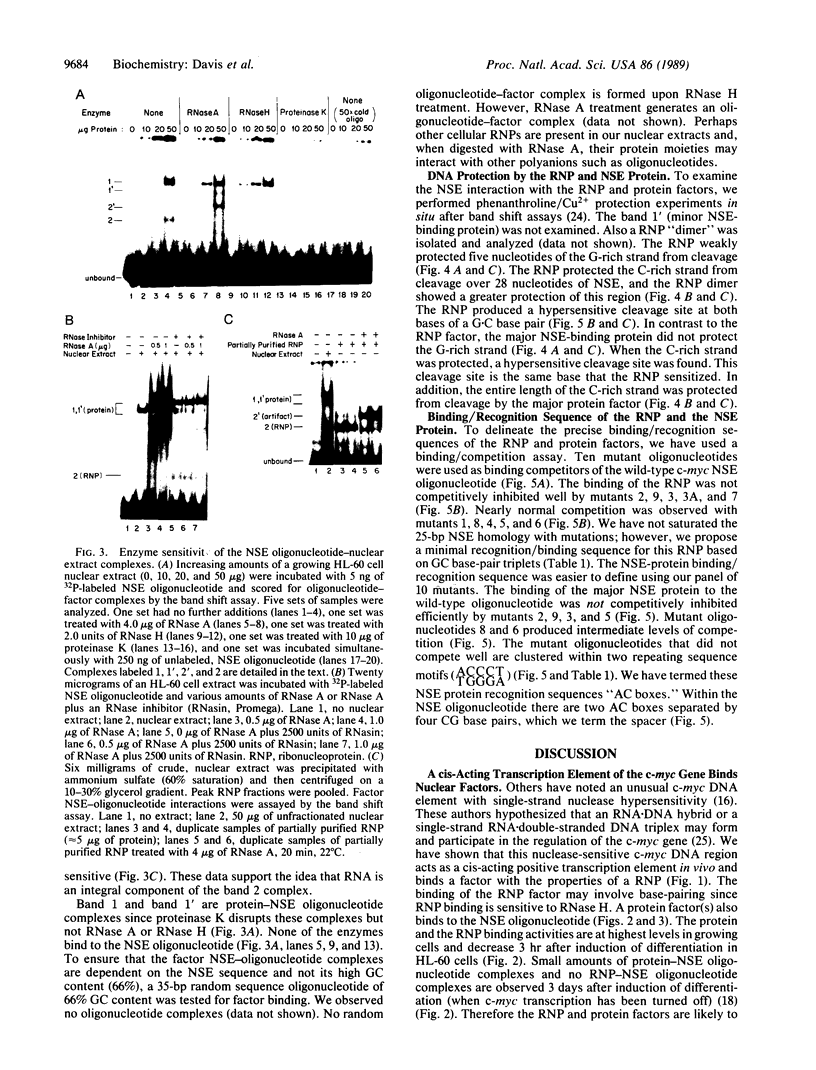
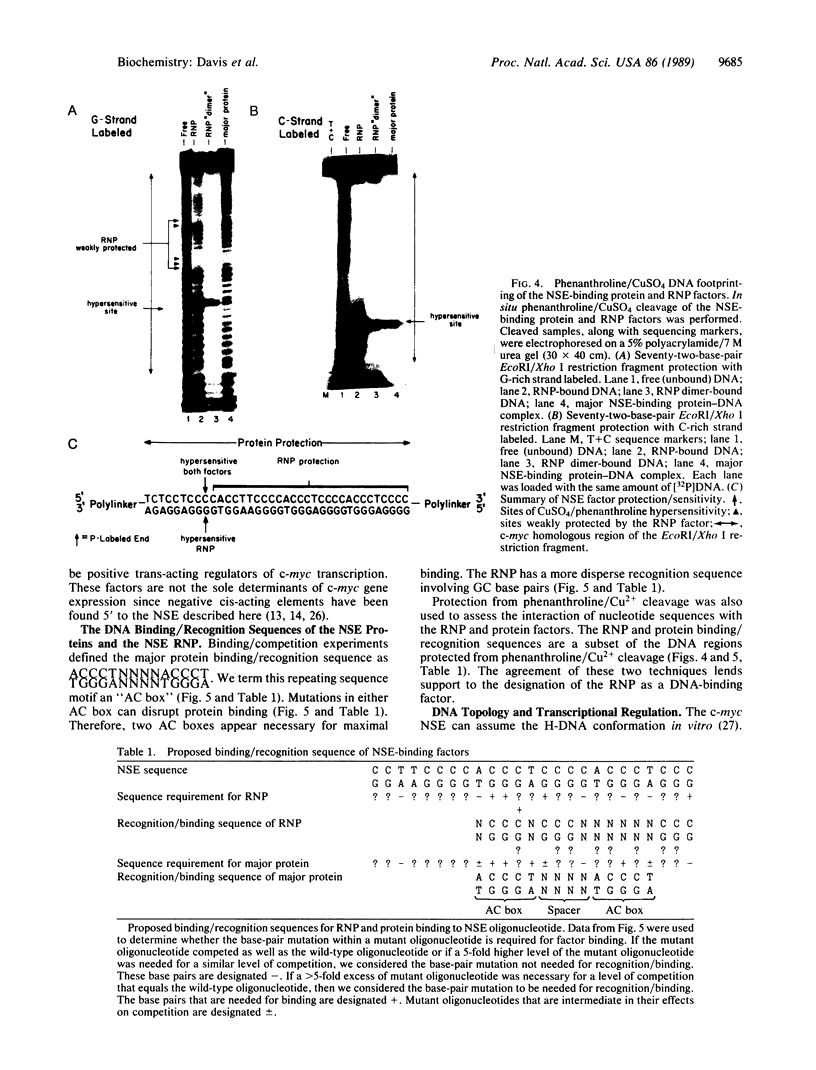
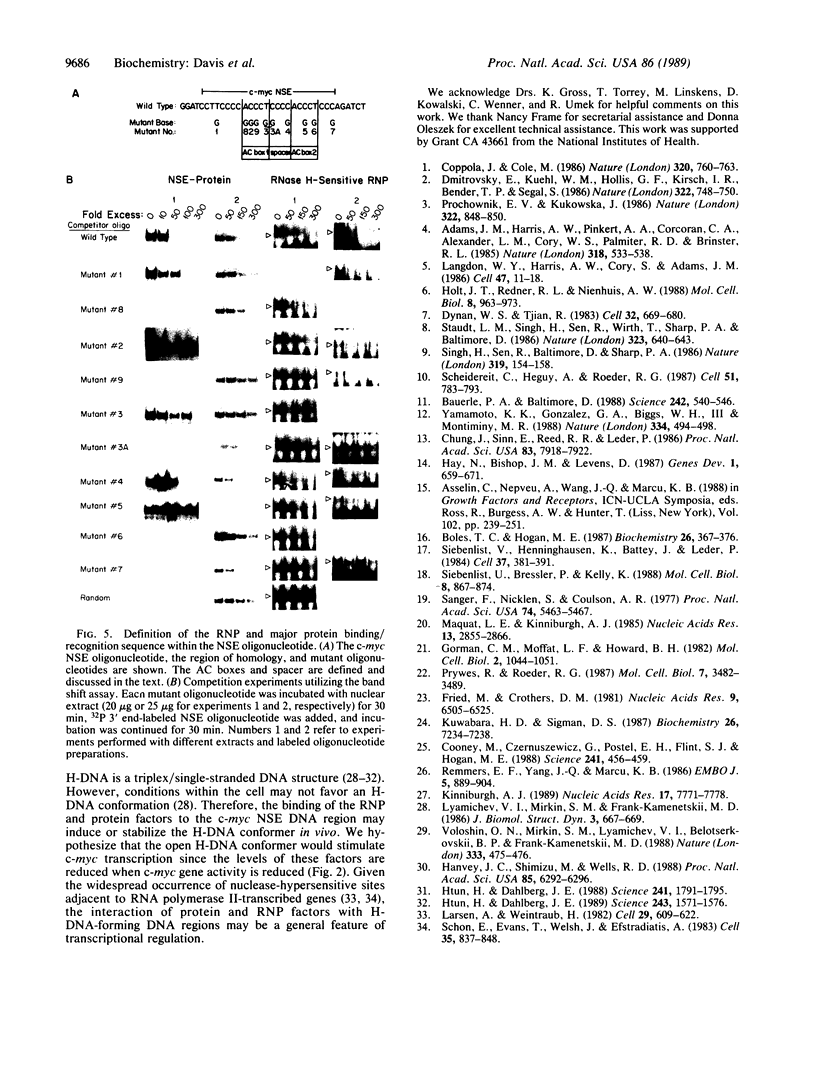
We have located a positive, cis-acting DNA sequence element within the 5' flanking DNA of the c-myc gene (-125 base pairs). This DNA sequence element has a large purine-pyrimidine strand asymmetry and can assume the H-DNA conformation. A factor with the properties of a ribonucleoprotein (RNP) interacts with this DNA region. The interaction of the c-myc DNA sequence element and the RNP involves an RNase H-sensitive mechanism and, therefore, may involve an RNA.DNA hybrid. In addition, a protein factor(s) binds to this DNA sequence element. DNA footprinting and mutant oligonucleotide binding/competition assays implicate a punctate, poly(G.C) recognition/binding sequence for the RNP factor, whereas the major protein factor requires two ACCCT sequence motifs for maximal binding. These results suggest that RNP and protein factors act as positive transcriptional regulators of the c-myc gene, perhaps by altering DNA topology.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Baeuerle P. A., Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988 Oct 28;242(4878):540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Boles T. C., Hogan M. E. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987 Jan 27;26(2):367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- Chung J., Sinn E., Reed R. R., Leder P. Trans-acting elements modulate expression of the human c-myc gene in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7918–7922. doi: 10.1073/pnas.83.20.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Holt J. T., Redner R. L., Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988 Feb;8(2):963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Kinniburgh A. J. A cis-acting transcription element of the c-myc gene can assume an H-DNA conformation. Nucleic Acids Res. 1989 Oct 11;17(19):7771–7778. doi: 10.1093/nar/17.19.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Langdon W. Y., Harris A. W., Cory S., Adams J. M. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986 Oct 10;47(1):11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J. A beta zero-thalassemic beta-globin RNA that is labile in bone marrow cells is relatively stable in HeLa cells. Nucleic Acids Res. 1985 Apr 25;13(8):2855–2867. doi: 10.1093/nar/13.8.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik E. V., Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. 1986 Aug 28-Sep 3Nature. 322(6082):848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- Prywes R., Roeder R. G. Purification of the c-fos enhancer-binding protein. Mol Cell Biol. 1987 Oct;7(10):3482–3489. doi: 10.1128/mcb.7.10.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Heguy A., Roeder R. G. Identification and purification of a human lymphoid-specific octamer-binding protein (OTF-2) that activates transcription of an immunoglobulin promoter in vitro. Cell. 1987 Dec 4;51(5):783–793. doi: 10.1016/0092-8674(87)90101-2. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. K., Gonzalez G. A., Biggs W. H., 3rd, Montminy M. R. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988 Aug 11;334(6182):494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]