Abstract
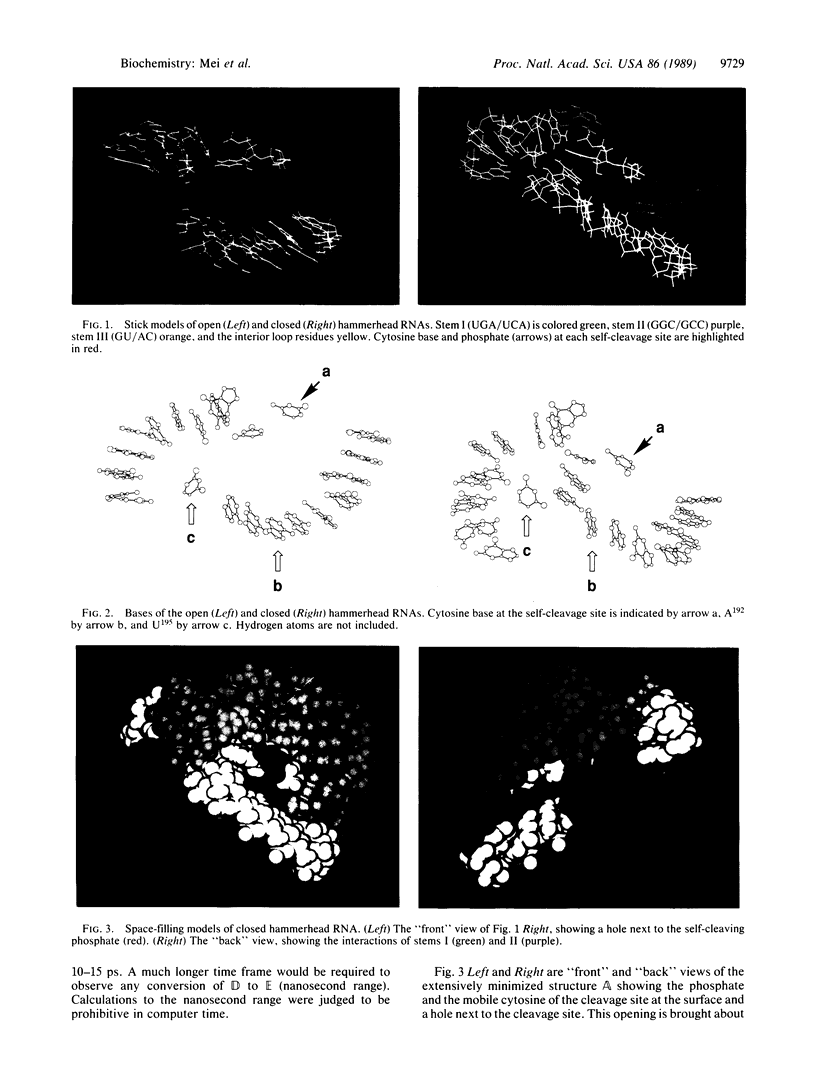
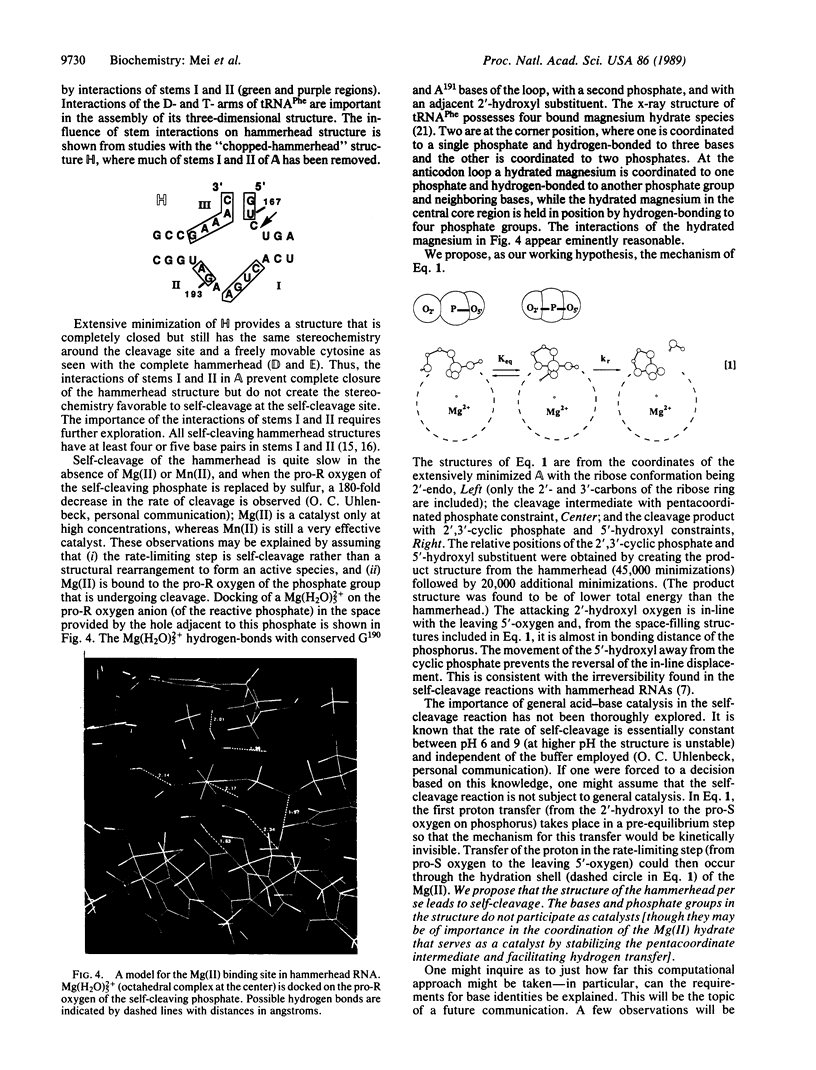
Extensive minimization and dynamics computational studies of the hammerhead structural domain of the virusoid of lucerne transient streak virus have been carried out. The following observations at the self-cleavage position are derived from the resulting three-dimensional structure: (i) the cytosine base is at the surface and does not interact with another base (it is free to move), and (ii) the ribose-phosphate backbone is forced to take an abrupt turn since it bridges stems I and III, and this turn points the pro-S and pro-R oxygens of the phosphate to the inward side of the hammerhead. These structural features are independent of the hammerhead being open or closed and allow an unencumbered 3'- to 2'-endo conformational change of the ribose with the resultant creation of an unusual stereochemistry that allows a direct in-line self-cleavage reaction. In the closed hammerhead structure, interactions of stems I and II create a vacancy into which the catalytic hydrated Mg(II) may be docked on labile phosphate. This opening is not present if stems I and II are shortened.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Doudna J. A., Gerber A. S., Cherry J. M., Szostak J. W. Genetic dissection of an RNA enzyme. Cold Spring Harb Symp Quant Biol. 1987;52:173–180. doi: 10.1101/sqb.1987.052.01.022. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Jeffries A. C., Sheldon C. C., Symons R. H. Structural and ionic requirements for self-cleavage of virusoid RNAs and trans self-cleavage of viroid RNA. Cold Spring Harb Symp Quant Biol. 1987;52:249–259. doi: 10.1101/sqb.1987.052.01.030. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries A. C., Symons R. H. A catalytic 13-mer ribozyme. Nucleic Acids Res. 1989 Feb 25;17(4):1371–1377. doi: 10.1093/nar/17.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. R. NMR studies of conformational states and dynamics of DNA. CRC Crit Rev Biochem. 1984;15(3):237–290. doi: 10.3109/10409238409102803. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Cleavage of specific sites of RNA by designed ribozymes. FEBS Lett. 1988 Nov 7;239(2):285–288. doi: 10.1016/0014-5793(88)80935-9. [DOI] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Levy G. C., Craik D. J., Kumar A., London R. E. A critical evaluation of models for complex molecular dynamics: application of NMR studies of double- and single-stranded DNA. Biopolymers. 1983 Dec;22(12):2703–2726. doi: 10.1002/bip.360221214. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Usher D. A. RNA double helix and the evolution of the 3',5' linkage. Nat New Biol. 1972 Feb 16;235(59):207–208. doi: 10.1038/newbio235207a0. [DOI] [PubMed] [Google Scholar]
- Usher D. A., Richardson D. I., Jr, Eckstein F. Absolute stereochemistry of the second step of ribonuclease action. Nature. 1970 Nov 14;228(5272):663–665. doi: 10.1038/228663a0. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]