Abstract
Duplications of distal 8p with and without significant clinical phenotypes have been reported and are often associated with an unusual degree of structural complexity. Here, we present a duplication of 8p23.1–8p23.2 ascertained in a child with speech delay and a diagnosis of ICD-10 autism. The same duplication was found in his mother who had epilepsy and learning problems. A combination of cytogenetic, FISH, microsatellite, MLPA and oaCGH analysis was used to show that the duplication extended over a minimum of 6.8 Mb between 3 539 893 and 10 323 426 bp. This interval contains 32 novel and 41 known genes, of which only microcephalin (MCPH1) is a plausible candidate gene for autism at present. The distal breakpoint of the duplicated region interrupts the CSMD1 gene in 8p23.2 and the medial breakpoint lies between the MSRA and RP1L1 genes in 8p23.1.
An interchromosomal insertion between a normal and polymorphically inverted chromosome 8 is proposed to explain the origin of this duplication. Further mapped imbalances of distal 8p are needed to determine whether the autistic component of the phenotype in this family results from the cumulative imbalance of many genes or dosage imbalance of an individual susceptibility gene.
Keywords: autism, epilepsy, duplication, 8p23.1–8p23.2, array CGH, MCPH1
Introduction
Chromosome 8 is an average chromosome with respect to length, gene content, repeat content and degree of segmental duplication.1 However, the final 15 Mb of the short arm is unusual in the variety of relatively complex chromosome anomalies that arise within it,2, 3, 4, 5, 6 the degree of copy number polymorphism associated with the defensin gene clusters7, 8, 9 and the degree of positive selection affecting the defensin and microcephalin (MCPH1) genes in humans.1
Clinically significant duplications of distal 8p can be distinguished from euchromatic variants5 but are still rare, and overlapping duplications are associated with marked variation in phenotype from severe10 to mild5, 6, 11 to unaffected.12, 13 Here, we present a new family with an interstitial duplication of distal 8p and a syndrome that includes autism in a child and epilepsy in the mother. The duplication has been mapped with fluorescence in situ hybridisation (FISH), microsatellites, multiplex ligation-dependent probe amplification (MLPA) and oligonucleotide array comparative genomic hybridisation (oaCGH). We briefly review overlapping duplications and the possible candidate genes for the autistic component of the phenotype.
Methods
Molecular cytogenetics
G-banded chromosomes were analysed at the 550 or higher band level and FISH carried out with 12 Ensembl tiling path (www.ensembl.org/homo_sapiens/cytoview) BACs, which map from bands 8p23.3 to 8p21.2 (Table 1). Genes assigned to the duplicated region were obtained from the Ensembl database (v32) using the MartView data export tool (www.ensembl.org/Multi/martview).
Table 1. BAC FISH, MLPA and oligonucleotide array results in mother and son.
| aBand | bBAC/microsatellite*/ MLPA**/array CGH loci*** | Start and stop position (Mb from telomere) | cResults |
|---|---|---|---|
| 8p23.3 | 338B22 | 467 644–668 409 | Normal |
| 8p23.3 | D8S504* | 1 004 963–1 005 161 | 136; 138 |
| 8p23.3 | D8S264* | 2 117 752–2 117 882 | 123; 127 |
| 8p23.2 | CSMD1 end | 2 782 789 | — |
| 8p23.2 | 336N16 | 2 888 584–3 078 882 | Normal |
| 8p23.2 | Oligo*** | 3 313 384–3 313 443 | Normal |
| 8p23.2 | D8S1824* | 3 539 893–3 540 122 | 227; 240; 246 |
| 8p23.2 | Oligo*** | 3 848 594–3 848 653 | Duplicated |
| 8p23.2 | 16H11 | 4 177 855–4 396 741 | Duplicated |
| 8p23.2 | D8S518* | 4 475 013–4 475 263 | 242; 244; 247 |
| 8p23.2 | CSMD1 start | 4 839 736 | — |
| 8p23.2 | RP5-991O23 | 5 316 020–5 469 277 | Duplicated |
| 8p23.1 | D8S1742* | 6 201 415–6 201 550 | 128;136;140 |
| 8p23.1 | CTD-2629I16 | 6 674 740–6 695 317 | Duplicated |
| 8p23.1 | DEFB1** | 6 715 511–6 722 939 | Duplicated |
| REPD | 122N11 | 7 367 548–7 578 862 | Three signals |
| 8p23.1 | 211C9 | 8 479 797–8 687 720 | Duplicated |
| 8p23.1 | MFHAS1 (MASL1)** | 8 680 942–8 787 978 | Duplicated |
| 8p23.1 | PPP1R3B | 9 032 916–9 045 616 | Duplicated |
| 8p23.1 | D8S503* | 9 270 573–9 270 784 | 244; 246; 250 |
| 8p23.1 | MSRA** | 9 949 240–10 323 808 | Duplicated |
| 8p23.1 | 112G9 | 10 028 624–10 236 504 | Duplicated |
| 8p23.1 | Oligo*** | 10 323 367–10 323 426 | Duplicated |
| 8p23.1 | Oligo*** | 10 511 353–10 511 405 | Normal |
| 8p23.1 | D8S520* | 10 593 772–10 593 964 | 188; 190 |
| 8p23.1 | Oligo*** | 10 660 074 | Normal |
| 8p23.1 | D8S550* | 10 918 965–10 919 233 | 256; 268 |
| 8p23.1 | GATA4** | 11 599 162–11 654 918 | Normal |
| 8p23.1 | 589N15 | 11 626 380–11 804 128 | Normal |
| REPP | — | — | — |
| 8p23.1 | D8S552* | 12 786 525–12 786 692 | 166; 174 |
| 8p22 | 433L7 | 14 277 096–14 462 154 | Normal |
| 8p22 | D8S1731* | 15 282 675–15 282 909 | 219; 233 |
| 8p22 | 809L8 | 18 239 259–18 460 085 | Normal |
| 8p21.3 | CGAT1** | 19 305 952–19 584 552 | Normal |
| 8p21.3 | D8S1786* | 22 489 342–22 489 551 | 207; 209 |
| 8p21.3 | D8S1771* | 25 497 152–25 497 377 | 227; 229 |
| 8p21.2 | 14I17 | 26 205 457–26 377 530 | Normal |
| 8q24.3 | 48I1 | 145 496 869–145 668 487 | Normal |
Grey shading indicates the extent of duplication; Italics used for CSMD1 start and endpoints.
G-dark bands in bold.
BACs RP11 unless indicated.
Numbers indicate the size of the microsatellite alleles found at informative loci.
Molecular genetics
DNA was extracted from whole blood using a salt precipitation technique. Fluorescent PCR amplification was carried out using standard conditions. Microsatellites were selected from within the duplicated interval using Ensembl and the Genome Database (www.gdb.org; Table 1). One primer from each pair was fluorescently labelled and the products analysed on an ABI 3100 automated sequencer.
MLPA was carried out using the standard protocol of MRC-Holland. Three additional in-house MLPA probes corresponding to the three genes on chromosome 8 (MFHAS1 (MASL1), DEFB1 and GATA4 – sequences are available upon request) were combined with MRC-Holland probe set P036, which contains five chromosome 8 probes (MFHAS1, PPP1R3B, MSRA, GATA4 and CGAT1).
Oligonucleotide array CGH
OaCGH was carried out using test genomic DNAs obtained using a standard salt extraction method and reference normal human male genomic DNA (Promega Corporation G147A 19813601, Madison, WI). DNA quantity and quality were assessed by UV/Vis spectrophotometry using the ND-1000 Spectrophotometer (NanoDrop Technologies, Rockland, DE) and gel analysis on a 0.8% agarose gel. The Agilent 44K Human Genome CGH microarray (G4410B, Batch number 0000013686, Agilent Technologies) consists of ∼43 000 60-mer oligonucleotide probes with an average spatial resolution of ∼35 kb and was applied to detect genome-wide copy number changes. In brief, patient and reference DNAs were digested with AluI and RsaI and the restriction enzymes were inactivated by incubation at 65°C. The Cy5- and Cy3-labelled DNA sample pair were combined and mixed with human Cot-1 DNA. Before hybridisation, the samples were heated at 95°C for 3 min and then incubated for 30 min at 37°C. Labelled target solution was hybridised to the 4 × 44K array with three other samples using SureHub chambers (G2534A) in a 65°C rotisserie oven (G2545A) set to rotate at 20 r.p.m. for 24 h. After hybridisation, the microarray slide was washed and dried according to the Agilent oligonucleotide array-based CGH for genomic DNA analysis protocol version 4.0 (G4140-90010). The microarray slide was scanned immediately using an Agilent microarray scanner (G2565BA) and image and data analyses were performed using the Agilent CGHAnalytics (v3.3) microarray software.
Clinical histories
The proband is the first-born child of unrelated parents of Argentinian origin. He was born after an otherwise uncomplicated pregnancy by normal delivery at term with a birth weight of 3.36 kg. Concerns about his development arose from around 2 years of age because of delayed speech. There had been mild delay in motor milestones with independent walking achieved at around 1.5 years. When assessed at the age of 3.5 years, he had around five single words in both English and Spanish but was described as having receptive language skills at a higher level. There was no jargon or echolalia or joint attention. He tended to play alone in nursery, but had no repetitive play. He would hand flap when excited and was upset by changes in routine. Eye-to-eye contact was poor and he demonstrated no spontaneous affection. He was diagnosed with autism satisfying the ICD-10 criteria. He had no seizures.
In addition to his developmental problems, there were complaints of poor feeding and diarrhoea. He required antitubercular treatment after developing hilar shadowing and a lower lobe collapse with a positive Mantoux test.
His development was monitored and had shown slow progress with persistent autistic features and continuing severe language delay. Examination showed a well-nourished non-dysmorphic boy; specifically, there were no features suggestive of Kabuki syndrome. His head circumference was above the 50th centile and height and weight on the 50th centile.
The proband's mother was aged 23 years at the time of his birth. She was diagnosed with epilepsy and learning problems. The degree of her difficulty was difficult to ascertain given her inability to speak English. She was non-dysmorphic on limited physical examination. She was prescribed two anticonvulsants before pregnancy, although she had discontinued them during pregnancy. Further details were uncertain; in addition, she was treated for tuberculosis during the pregnancy. The proband's father, aged 33 years at the time of his birth, had been diagnosed with psychiatric problems and had a previous history of substance abuse; details of his medical condition and treatment are unknown.
The couple have a second son 3 years younger than the proband. At the age of 1.5 years, this boy has mild expressive language delay but no features in the autistic spectrum. He is non-dysmorphic and has normal growth parameters.
Results
G-banded analysis showed a duplication of the distal short arm of chromosome 8 in the proband and his mother (Figure 1A–B) and normal chromosomes in the proband's father and younger brother. Using dual colour FISH in both mother and son, five BACs from bands 8p23.1 and 8p23.2 were duplicated (Figure 2 a–d) and BAC RP11-122N11 gave three signals (Figure 2b; Table 1). All other probes gave normal results (Table 1). FISH and molecular genetic analysis, using microsatellite and MLPA probes, indicated that the duplication of 8p23.1–8p23.2 includes a minimum of ∼6.7Mb between 3 539 893 (D8S1824) in 8p23.2 and 10 236 504 bp (112G9) in 8p23.1 (Table 1 based on Ensembl v36/7).
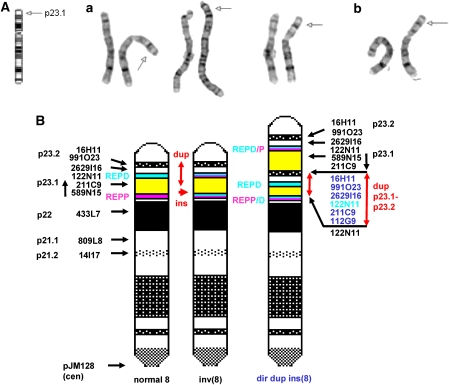
Figure 1.
(A and B) Partial karyotypes of (a) the proband and (b) his mother. Note the increased interval between bands p22 and p23.2 and the additional G-dark in the proximal part of the increased interval (arrowed on the right hand duplicated chromosome in each case). (B) Idiogram of the proposed insertional duplication of distal 8p23.1–8p23.2 from a normal chromosome 8 into a polymorphically inverted chromosome 8 adjacent to REPP/D in proximal 8p23.1. On the left hand normal chromosome, distal 8p23.1 is in white, REPD is in blue, REPP is in purple, the interval between them in yellow and proximal 8p23.1 in white again. The double-headed vertical red arrow indicates the extent of the duplication on the normal chromosome and the vertical black upward arrow next to BACs RP11-211C9 and RP11-589N15 indicates the ‘genome browser' orientation of the REPP to REPD interval. In the central chromosome, REPP and REPD are shown in both colours as the result of the common polymorphic inversion and the horizontal single-headed red arrow indicates the proposed point of insertion adjacent to the composite REPD/P repeat. The duplicated chromosome is on the right with the vertical double-headed red arrows showing the position and extent of the duplication. Duplicated BACs are shown in blue and the BAC, which gave three signals (RP11-122N11) is in turquoise. Note the orientation of the duplicated BACs is the same as that on the left hand normal chromosome. By contrast, the reverse order of BACs RP11-589N15 and RP11-211C9 is indicated by the downward vertical black arrow.
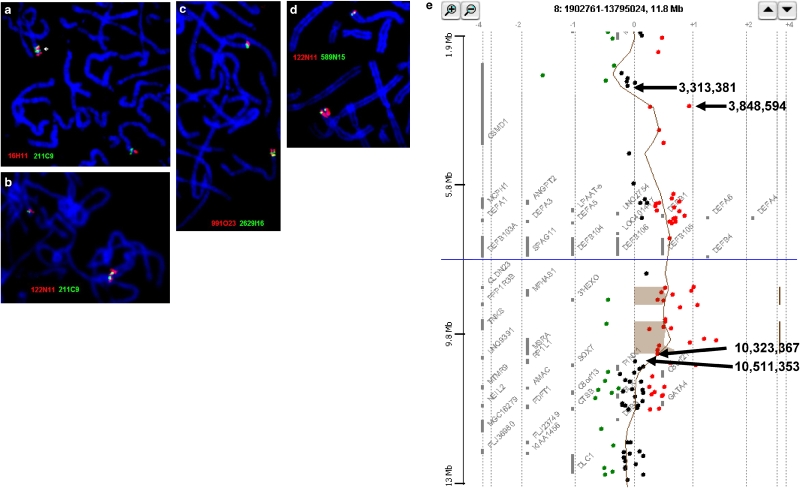
Figure 2.
(a–d) Representative dual colour FISH in metaphases from the proband and mother; (a) direct duplication of both RP11-16H11 (red) from 8p23.2 and RP11-211C9 from 8p23.1 (green) in the proband; (b) the duplication of RP11-211C9 (green) alternating between three distinct RP11-122N11 signals (red) in a higher resolution cell in the mother; (c) direct duplication of both RP5-991O23 (red) from 8p23.2 and CTD-2629I16 from 8p23.1 (green) in the mother; (d) the single-copy RP11-589N15 signals (green) located very distally within the block of signals from RP11-122N11 in the mother. (e) Agilent 44K oligonucleotide array result from the proband analysed with Analytics v3.3 software. The black arrows point to the normal and duplicated clones that flank the duplication with the base pairs given for the p-telomeric edge of each oligonucleotide probe (see also Table 1).
OaCGH was used to look for further possible imbalances in 8p or at other sites across the genome and confirmed a duplication with a minimum size of 6.47 Mb between 3 848 594 and 10 323 426 bp (Ensembl v36/7; Figure 2e; Table 1). The array result narrowed the distal duplication breakpoint to 226 kb between the duplicated microsatellite locus D8S1824 at 3 539 893 bp and the first normal oligonucleotide clone distal to the duplication at 3 313 443 bp. This interval extends from introns 7 to 11 of CSMD1 and must therefore interrupt this gene. The array results place the medial 8p23.1 breakpoint within the 188 kb between the last duplicated oligonucleotide at 10 323 426 bp and the first normal oligonucleotide at 10 511 353 bp. When the array and informative microsatellite results are combined, the duplication is a minimum of 6.78 Mb in size between 3 539 893 and 10 323 426 bp and a maximum of 7.19 Mb between 3 313 443 and 10 511 353 bp (Table 1).
Two further possible small areas of duplication with log2 ratios of 0.38 were identified; 16 clones covering 231 kb between 145 464 363 and 145 695 514 bp in 8q24.3 and 11 clones covering 194 kb between 48 448 631 and 48 431 311 bp in 3p21.31. A duplication within 8q24.3 was not confirmed using BAC RP11-48I1 and there were no suitable FISH clones with which to test the 3p region, which corresponds to the Plexin-B1 precursor gene that has no known pathology (OMIM 601053).
The duplication is thought to be insertional because of the closeness of the G-dark duplicated band 8p23.2 to G-dark 8p22 (Figure 1A–B), the three signals from REPD BAC RP11-122N11 (Figure 2b and d), the very distal location of the single-copy BAC 589N15 on the duplicated chromosome (Figure 2d), and the three alleles that were seen with five independent microsatellite markers (Table 1).
The karyotype of the proband can therefore be written as:
46,XY,dir dup ins(8)(p23.1;p23.1p23.2)mat.ish dir dup ins(8)(pter->p23.1::p23.2->p23.1::p23.1->qter) (RP11-338B22+,RP11-336N16+,RP11-16H11+,RP5-991O23+,CTD-2629I16+,RP11-122N11+,RP11-589N15+,RP11-112G9+,RP11-211C9+,RP11-16H11+,RP5-991O23+,CTD-2629I16+,RP11-122N11+,RP11-211C9+,RP11-112G9+,RP11-122N11+,RP11-433L7+,RP11-809L8+,RP11-14I17+,RP11-48I1+).arr cgh dup(8)(p23.1p23.2)(B35:CHR8:3,848,594->10,323,426++).
Discussion
This family has a directly transmitted duplication of 8p23.1-p23.2 with a minimum size of 6.78 Mb extending from 3 539 893 to 10 323 426 bp (shaded area Table 1) and a phenotype that includes speech delay, autism, epilepsy and learning difficulties. This is apparently a unique rearrangement as we are not aware of a direct precedent for this duplication in the literature and there was no evidence for predisposing repeats at the duplication breakpoints of the kind associated with other recurrent complex rearrangements of 8p.2, 3, 6, 10
The medial 8p23.1 breakpoint of the inserted region is unusual in that it occurs between rather than within the olfactory receptor repeats (REPD and REPP) that map to both ends of band 8p23.1 and mediate other 8p rearrangements.2 The duplication is direct (Figure 2a) but is thought to be insertional because:
The duplicated part of G-dark 8p23.2 is quite close to band 8p22 in the partial G-banded karyotypes (Figure 1A) and therefore difficult to reconcile with a simple direct duplication.
There are three signals from the REPD BAC RP11-122N11 (Figure 2b), which contains variable defensin gene cluster sequences. The most distal and proximal signals are located on the chromosome, which received the insertion. The signal between them is located on the inserted segment. The possible triplication of the variable defensin domain was not confirmed by oaCGH but this is likely to have been a reflection of the use of control DNA of unknown defensin copy number.
BAC RP11-589N15 is normally proximal to BACRP11-211C9 but was clearly distal to the duplication and very close to the most distal signal from REPD BAC RP11-122N11 (Figure 2d). This suggests that the duplicated segment was inserted into a chromosome with the polymorphic inversion of 8p23.1 in which the order of RP11-211C9 and RP11-589N15 was reversed (Figure 1B).
Three alleles were seen with five independent microsatellite markers (Table 1).
We also considered that the duplication might have been inserted into one of the repeats but this would have resulted in four signals – one from the repeat unaffected by the insertion, one from the repeat contained within the duplication and two signals from the repeat split by the insertion. This was not observed and our best interpretation was therefore an insertion adjacent to a composite REPP/D as illustrated in Figure 1B.
The 8p23.1 breakpoint and duplication in this family creates an additional truncated copy of the CUB and sushi multiple domains gene CSMD1 (OMIM 608397) from the 5′ prime end to a breakpoint between intron 7 at 3 313 384 bp and intron 11 at 3 539 893 bp in band 8p23.2. However, this is unlikely to be pathogenic as two copies of the normal gene remain and copy number variation including truncated copies of this gene have been reported extending to 1.3 Mb (L1013 in the Database of Genomic Variants)14 and 2.3 Mb in size (cnp676).15 The medial 8p23.1 breakpoint of the inserted segment, lies between the duplicated peptide methionine sulfoxide reductase A (MSRA) and the retinitis pigmentosa 1-like protein 1 (RP1L1) gene outside the duplication. In animal models, the overexpression of MSRA is protective against oxidative damage (OMIM 601250) and no pathology has yet been associated with RPL1L1 (OMIM 608581). The insertion site is adjacent and proximal to the proximal REPP/D in the inverted chromosome. Thus, the breakpoints do not harbour strong candidates for the phenotype in this family.
The duplication in this family includes 41 known and 32 novel genes. However, as many as 22 of the known genes are members of the α- and β-defensin cluster regions, which vary in copy number in the normal population7, 9 and are therefore unlikely to be responsible for the phenotype. Of the remaining 19 genes, few are obvious candidates for the phenotype in this family. However, the 8p23.1 duplication syndrome mediated by REPP and REPD is associated with mild dysmorphism, speech delay and learning difficulties but not yet with autism.5, 6 If this region is excluded together with the defensin gene clusters, only five known genes remain (CSMD1, MCPH1, ANGPT2, AGPAT5 and NP_997295) of which only MCPH1 has an OMIM morbid entry. Recessive mutations and deletions of this gene (OMIM 251200, 606858 and 607117) are associated with microcephaly, mental retardation and premature chromosome condensation16 and additional copies of this gene have been associated with the increased head size found in two previously reported patients with 8p duplications and triplications by Giorda et al.10 As there is increasing evidence of an association between rapid head growth in the first year of life and subsequent autism,17, 18 it is conceivable that the duplication of this gene could give rise to autism or less severe psychomotor developmental problems in patients with additional copies of MCPH1.
Other duplications of 8p23.1 have been reported with a wide variety of presentations, including developmental delay, heart disease and autism but their gene content has not been determined yet.19, 20 A cytogenetic duplication of 8p21 to 8p23 was also associated with autism21 but, like the well-known inverted duplications of 8p (inv dup del(8)s), includes a large more proximal segment of 8p, which contains other candidate genes for syndromic autism.
Our results add to the extreme heterogeneity found in both syndromic and non-syndromic autism using array CGH.22, 23, 24 In particular, Jacquemont et al.22 found nine different imbalances in 29 patients with syndromic autism, only one of which corresponded to a known autism susceptibility locus. In addition, Sebat et al.23 found distinct gains and losses in another 17 patients but none of these 26 imbalances or de novo copy number variants involved 8p. However, the recently established Autism Chromosome Rearrangement database contains a variety of cytogenetic rearrangements that suggest that the distal 8p may yet harbour one or more autism susceptibility loci.24
In conclusion, we report a duplication of 8p23.1 to 8p23.2 associated with speech delay, autism, epilepsy and learning difficulties. At the current level of knowledge, only MCPH1 is a plausible candidate for the autistic component of the phenotype in this family. Thus, further mapped imbalances of distal 8p are needed to provide better prognostic information for patients in the future and to investigate whether the autism results from the cumulative imbalance of many of the genes involved or dosage imbalance of an individual susceptibility gene.
URLs
Ensembl tiling path (http://www.ensembl.org/homo_sapiens/cytoview)
MartView data export tool (www.ensembl.org/Multi/martview).
OMIM (http://www.ncbi.nlm.nih.gov/entrez/query)
Database of Genomic Variants (http://projects.tcag.ca/variation/)
The Autism Chromosome Rearrangement Database (http://projects.tcag.ca/autism/)
Acknowledgments
We are grateful to the Sanger Centre for providing the Ensembl tiling path clones and to Dr Hiroaki Shizuya for kindly providing BAC 51D11. SH and VKM are supported as part of the National Genetics Reference Laboratory (Wessex) by the UK Department of Health Genetics, Embryology and Assisted Conception Unit. The image enhancement equipment used for this work was provided by the Welcome Trust and Trust Funds of Salisbury NHS Health Care Trust
References
- Nusbaum C, Mikkelsen TS, Zody MC, et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H, Harada N, Ida T, et al. Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics. 2003;82:238–244. doi: 10.1016/s0888-7543(03)00108-3. [DOI] [PubMed] [Google Scholar]
- Barber JCK, Maloney V, Hollox EJ, et al. Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet. 2005;13:1131–1136. doi: 10.1038/sj.ejhg.5201475. [DOI] [PubMed] [Google Scholar]
- Barber JCK, Maloney VK, Huang S, et al. 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet. 2008;16:18–27. doi: 10.1038/sj.ejhg.5201932. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a β-defensin antimicrobial gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudien S, Galgoczy P, Huse K, et al. Polymorphic segmental duplications at 8p23.1 challenge the determination of individual defensin gene repertoires and the assembly of a contiguous human reference sequence. BMC Genomics. 2004;5:92–103. doi: 10.1186/1471-2164-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred PM, Hollox EJ, Armour JAL. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum Mol Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- Giorda R, Ciccone R, Gimelli G, et al. Two classes of low-copy repeats co-mediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat. 2007;28:459–468. doi: 10.1002/humu.20465. [DOI] [PubMed] [Google Scholar]
- Brooks SS, Genovese M, Gu H, Duncan CJ, Shanske A, Jenkins EC. Normal adaptive function with learning disability in duplication 8p including p22. Am J Med Genet. 1998;78:114–117. doi: 10.1002/(sici)1096-8628(19980630)78:2<114::aid-ajmg3>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- Engelen JJM, Moog U, Evers JLH, Dassen H, Albrechts JCM, Hamers AJH. Duplication of chromosome region 8p23.1->p23.3: a benign variant. Am J Med Genet. 2000;91:18–21. [PubMed] [Google Scholar]
- Harada N, Takano J, Kondoh T, et al. Duplication of 8p23.2 a benign: cytogenetic variant. Am J Med Genet. 2002;111:285–288. doi: 10.1002/ajmg.10584. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003;290:337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- Dawson G, Munson J, Webb SJ, Nalty T, Abbott R, Toth K. Rate of head growth decelerates and symptoms worsen in the second year of life in autism. Biol Psychiatry. 2007;61:458–464. doi: 10.1016/j.biopsych.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-H, Graw SL, McGavran L. 8p23 duplication reconsidered: is it a true euchromatic variant with no clinical manifestation. J Med Genet. 2002;39:769–774. doi: 10.1136/jmg.39.10.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SJ, Teebi AS, Adatia I, Teshima I.Inherited duplication, dup(8)(p23.1p23.1) pat, in a father and daughter with congenital heart defects Am J Med Genet 200110479–80.(letter). [DOI] [PubMed] [Google Scholar]
- Papanikolaou K, Paliokosta E, Gyftodimou J, et al. A case of partial trisomy of chromosome 8p associated with autism. J Autism Dev Disord. 2006;36:705–709. doi: 10.1007/s10803-006-0104-3. [DOI] [PubMed] [Google Scholar]
- Jacquemont ML, Sanlaville D, Redon R, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Genome Consortium Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]