Abstract
Paracentric inversions (PAIs) are structural chromosomal rearrangements generally considered to be harmless. To date, only a few studies have been performed concerning the meiotic segregation of these rearrangements, using either the human–hamster fertilization system or fluorescence in situ hybridization (FISH) with centromeric or telomeric DNA probes. To improve the assessment of imbalances in PAI, we present a new strategy based on FISH assay using multiple bacterial artificial chromosome probes, which allow a precise localization of chromosome break points and the identification of all meiotic products in human sperm. Sperm samples of three cases with PAI were investigated: an inv(5)(q13.2q33.1), an inv(9)(q21.2q34.13) and an inv(14)(q23.2q32.13). The frequencies of spermatozoa with inverted chromosomes were 44.7% in inv(5), 42.7% in inv(9) and 46.7% in inv(14). The global incidences of unbalanced complements were 9.7, 12.6 and 3.7% in inv(5), inv(9) and inv(14), respectively. This report is the first study providing a detailed description of meiotic segregation patterns in human sperm by using a sperm FISH approach. This study demonstrates that the detailed analysis of segregation in PAI may provide important data for both genetic analysis and counseling of inversion carriers.
Keywords: paracentric inversion, meiotic segregation analysis, sperm FISH
Introduction
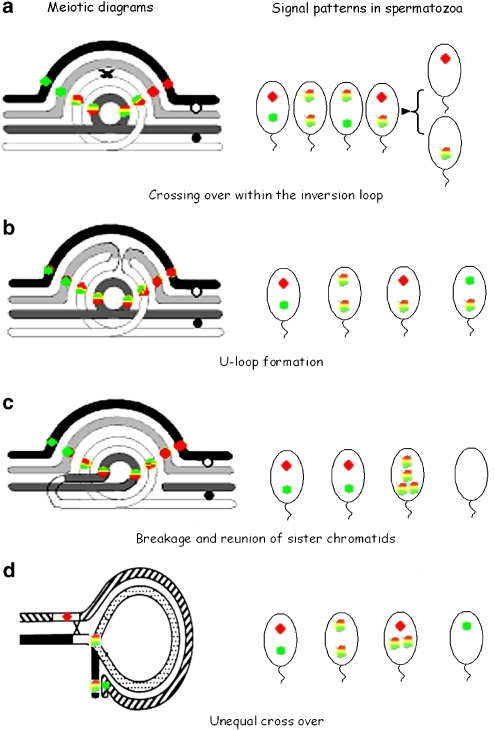
Paracentric inversions (PAIs) are chromosomal rearrangements that occur after two breaks in a chromosome arm followed by rotation of the chromosomal segment through 180° and reinsertion of the segment. The presence of PAI may be unnoticed as they do not alter the chromosome arm ratio. Consequently, it is suspected that a certain percentage of small PAI remains undetected.1 Estimates of frequency range from 0.1 to 0.5%2, 3 and PAIs are generally considered to be harmless for the carrier. Theoretically, heterozygous carriers of a PAI cannot produce viable unbalanced progeny. During meiosis, the occurrence of one (or an uneven number of) crossing-over event(s) within the inversion loop of affected segments should produce one dicentric and one acentric recombinant chromosomes, both of which are considered to be lethal. However, several cases of viable recombinant offsprings are reported,3, 4, 5, 6 and a variety of mechanisms was proposed for the meiotic formation of stable recombinant chromosomes with deletion and/or duplication, including breakage of dicentric recombinants,1 breakage and reunion of sister chromatids,7 unequal crossover8 and the abnormal process of U-loop recombination.9, 10 Also, the occurrence of spontaneous abortion, recurrent miscarriage and infertility refute complete innocuousness of the parental PAI and suggests that recombination here could arise more frequently than suspected from liveborn data.
The incidence of viable recombinants has been estimated to be 3.8%11 but this figure remains controversial because of ascertainment biases and incomplete data.12, 13 The direct chromosomal analysis of gametes allows overcoming this lack of information. However, only very few segregation analysis studies were performed for PAI.
Cheng et al14 analyzed a PAI of chromosome 7 in human oocytes and found a classical inversion loop in 10% of cells. Sperm karyotyping using the human sperm/hamster oocyte method was performed on two PAIs, one with inv(7)(q11q22) and the other with an inv(14)(q24.1q32.1). Brown et al15 used the sperm typing technique for studying patterns of recombination in sperm from a heterozygous carrier of a PAI of chromosome 9. Both these procedures were labor-intensive and provided data on small numbers of spermatozoa (from 94 to 282). The fluorescence in situ hybridization (FISH) technique has offered a new approach for segregation study in human sperm. However, only four PAIs have been investigated to date using the sperm FISH procedure with centromeric and/or telomeric DNA probes. These analyses provided data on large samples of human sperm (from 496 to 8158), but they did not allow the in situ identification of all the meiotic segregates because of the incomplete hybridization patterns obtained with centromeric or telomeric DNA probes.
To precisely determine the segregation patterns of chromosomal inversions in human gametes, we developed a high-resolution mapping strategy, based on the use of bacterial artificial chromosome (BAC) clones spanning the chromosomal break points. In the present study, we report the first application of this in situ high-resolution procedure on human sperm samples, for the direct segregation analysis of an inv(5)(q13.2q33.1), an inv(9)(q21.2q34.13) and an inv(14)(q23.2q32.13).
Materials and methods
Three otherwise healthy men heterozygous for PAI inv(5), inv(9) and inv(14) were studied by molecular cytogenetics. The three patients, aged 60, 44 and 30 years respectively, were all ascertained through a fertility workup. All of them displayed normal sexual development and normal semen parameters (sperm count >40 million/ml; >50% normal morphology and >50% progressive motility). The three subjects were informed of the investigations planned, and they signed a consent form before participation in this study, which was approved by the ethical board of Montpellier University Hospital.
Initial cytogenetic studies of inv(5) and inv(14) were carried out on peripheral blood lymphocytes from the inversion carriers, using standard cytogenetic procedures for R- and G-banding. The chromosomal preparations from both the normal subjects and the inversion carriers were used for testing BAC probes and optimizing the in situ hybridization conditions. The selection of BAC clones and their preparation were performed as previously described.16 The breakpoint spanning BACs (Table 1) were used in sperm FISH along with a control BAC located outside the breakpoint regions. Sperm samples were processed according to the procedure of Pellestor et al.16 Briefly, each sample was washed three times in 1 × PBS by centrifugation (300 g, 5 min) and fixed for 1 h in fresh fixative (3:1 methanol:glacial acetic acid) at –20°C. The sperm suspension was then dropped onto clean microscope slides and air-dried. Slides were aged 2 days at room temperature before use for in situ chromosomal labeling. Before FISH procedure, the slides were immersed for 10 min in a pepsin solution (50 ng/ml in 0.01 HCl) prewarmed at 37°C, washed for 2 min in 1 × PBS and then dehydrated through an ethanol series (70, 90 and 100%) and air-dried. The sperm nucleus decondensation and DNA denaturation was performed by slide incubation in 0.5 NaOH solution at room temperature for 8 min, followed by a wash in 2 × SSC, dehydration through an ethanol series and immersion in 70% formamide/2 × SSC solution 3 min at 73°C. Finally, the slides were washed in 2 × SSC, dehydrated through an ethanol series and air-dried.
Table 1. BAC clones used for segregation analysis.
| Cytogenetic region | BAC/PAC clones | Position |
|---|---|---|
| 5q13.3 | RP11-114B1 | 74827703–74992978 |
| RP11-9N21 | 74962159–75118483 | |
| RP11-179H5 | 74998605–75170876 | |
| RP11-205H11 | 75138142–75273622 | |
| 5q33.1 | RP11-263K13 | 150080197–150239374 |
| RP11-12A4 | 150265542–150418860 | |
| RP11-96P14 | 150474047–150660861 | |
| 9q21.2 | RP11-267G14 | 79046423–79213265 |
| RP11-466A17 | 79213269–79372174 | |
| RP11-844C21 | 79301878–79514928 | |
| RP11-235F20 | 132993351–133152079 | |
| RP11-105C7 | 132816295–132908901 | |
| 9q34.13 | RP11-544A12 | 132955072–133152093 |
| RP11-643E14 | 133146456–133324218 | |
| 14q23.2 | RP11-676P5 | 63629431–63768477 |
| RP11-712C19 | 63768477–63912834 | |
| 14q32.13 | RP11-262P9 | 93946936–94127939 |
| RP11-986E7 | 94090988–94303514 | |
| RP11-179A9 | 94303515–94457888 |
The breakpoint characterization of inv(5) and inv(14) as well as the use of the breakpoint spanning BACs in sperm FISH analysis was published elsewhere.17 In the case of inv(9), only sperm sample was available. The BACs were selected according to the break points determined cytogenetically. A cocktail of continuous BAC clones was labeled with either digoxigenin-11-dUTP or biotin-16-dUTP. Sperm FISH was performed to narrow down the breakpoint region. The BACs in contiguity spanning the break points (Table 1) were labeled and used as FISH probes.
Results
The labeling efficiency of BAC probes was determined on lymphocyte preparations and sperm preparations. The hybridization of the selected clones gave efficient results on both types of preparations with a satisfactory visualization of fluorescent signals as illustrated in Figure 1. For each probe mixture, the hybridization efficiency was estimated by scoring 50 metaphases and 100 sperm nuclei. The efficiency values ranged from 99.5 to 100% according to the probe set.
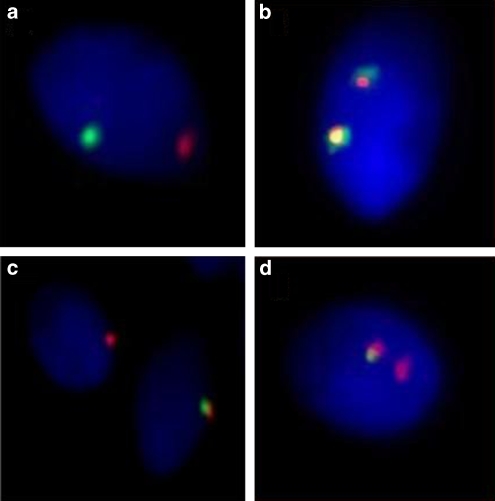
Figure 1.
Meiotic diagrams and FISH signal patterns in spermatozoa using breakpoint-specific BAC clones. When using specific BAC clones spanning the break points, the in situ mixing of green and red signals gives yellow signals or a combined red/green signal on inverted or recombinant chromosomes, and thus allows the identification of each meiotic product, like (a) normal, (b) inverted, (c) duplicated/deficient, and (d) dicentric.
The size of the PAI in chromosome 5 was estimated as 41% of the total length of chromosome 5. The BACs spanning the break points of 5q13.3 and 5q33.1 regions were RP11-179H5 and RP11-12A04, respectively. As shown in Figure 1a, the BACs RP11-179H5 and RP11-12A04 gave clear green and red signals, respectively, in normal spermatozoa.
The size of inversion in chromosome 9 was 39% of the total chromosome length. The break points at 9q21.2 and 9q34.13 were narrowed down to BAC contiguities RP11-267G14/RP11-466A17/RP11-844C21 and RP11-105C7/RP11-235F20/RP11-544A12/RP11-643E14, respectively. In sperm harboring the inverted chromosome (Figure 1b), BAC probes gave specific patterns with either two yellow signals or two combined red and green signals, corresponding to the perfect or the incomplete colocation of BAC clones. On the other hand, sperm with a duplicated/deficient chromosome gave either one red or one combined red/green signal, as illustrated in Figure 1c. Sperm nuclei with a dicentric chromosome displayed one red signal and one combined red/green signal (Figure 1d).
The size of the inverted segment in chromosome 14 was 29% of the total chromosome length. BACs spanning the break points of 14q23.2 and 14q32.13 regions were RP11-712C19 and RP11-986E7, respectively.
A total of 4807, 1608 and 7670 sperm nuclei were evaluated for inv(5), inv(9) and inv(14), respectively. The proportion of inverted chromosomes was 44.7% in inv(5), 42.7% in inv(9) and 46.7% in inv(14). The incidence of recombinants was 9.7% for inv(5), 12.6% for inv(9) and 3.7% for inv(14). Details of segregation analysis of these three inversions as well as of seven other PAIs previously analyzed are summarized in Table 2.
Table 2. Results of segregation analysis in paracentric inversions using sperm karyotype, sperm typing, sperm FISH or breakpoint FISH.
| Inversion | No. of sperms counted | No. of sperms with normal chromosome (%) | No. of sperms with inverted chromosome (%) | No. of sperms with duplicated/ deficient chromosome (%) | No. of sperms with dicentric chromsome (%) | % Recombinant | Inverted segment size in Mb (%)a | Method | Author |
|---|---|---|---|---|---|---|---|---|---|
| inv(5)(q13.3q33.1) | 4807 | 2194 (45.6) | 2147 (44.7) | 420 (8.7) | 46 (1) | 9.7 | 75 (41) | Breakpoint FISH | This study |
| inv(9)(q21.2q34.13) | 1608 | 719 (44.7) | 687 (42.7) | 181 (11.3) | 21 (1.3) | 12.6 | 54 (39) | Breakpoint FISH | This study |
| inv(14)(q23.2q32.13) | 7670 | 3806 (49.6) | 3579 (46.7) | 263 (3.4) | 22 (0.3) | 3.7 | 30 (29) | Breakpoint FISH | This study |
| inv(7)(q11q22) | 94 | 34 (36) | 59 (63) | 0 | 0 | 0 | 50 (32) | Sperm karyotype | Martin18 |
| inv(9)(q32q34.3) | 282 | ? | ? | 0 | 0 | 0 | 32 (24) | Sperm typing | Brown et al15 |
| inv(14)(q24.1q32.1) | 120 | 50 (42) | 70 (58) | 0 | 0 | 0 | 27 (26) | Sperm karyotype | Martin19 |
| inv(2)(q14.2q24.3) | 496 | ? | ? | ? | 4 (0.8) | 0.8 | 49 (20) | Sperm FISH | Devine et al20 |
| inv(4)(p14p15.3) | 8158 | ? | ? | ? | ? | 0.03 | 11 (5) | Sperm FISH | Anton et al21 |
| inv(11)(q13.2q14.3) | 1001 | ? | ? | ? | 4 (0.4) | 0.4 | 27 (20) | Sperm FISH | Vialard et al22 |
| inv(12)(q15q24.1) | 1000 | ? | ? | ? | 5 (0.5) | 0.5 | 44 (33) | Sperm FISH | Vialard et al22 |
Inverted segment size in percentage is calculated for the entire chromosome
Data from previous studies are shown in italic characters.
Discussion
The meiotic behavior of chromosomal PAI has hardly been explored in man. This may be linked to the low risk of imbalance generally attributed to chromosomal inversions. However, as indicated in Table 2, the technical difficulties inherent to the precise identification of all meiotic products in PAI also constitute important limitations for accurate segregation analysis in previous studies. Both sperm typing assay and sperm FISH with centromeric or telomeric probes were unable to identify all possible inversion segregants. Here, we demonstrate that BAC clones spanning the break points can be efficiently used as probes for precisely determining the segregation pattern of chromosomal inversions in human sperm. Of course, such an in situ mapping of break points is labor-intensive and takes longer than the use of commercial centromeric and subtelomeric probes. In addition, several authors have pointed out the lack of efficiency of subtelomeric probes for the identification of recombinant chromosomes in human sperm, and consequently the possible erroneous estimate of imbalance risk in male inversion carriers.23, 24, 25 However, by application of BAC probes, a complete and precise segregation analysis can be carried out, whereas previous sperm FISH studies have reported approximate rates of imbalances, with no details on dicentric and acentric recombinant products (Table 2). The present approach allows to distinguish all possible segregants from each recombination mechanism described in PAI (Figure 2), and thus to exclude the possibility of missing ones. These detailed data might contribute to a better exploration of the meiotic behavior of PAI, and they might also significantly improve the genetic counseling provided to inversion carriers.
Figure 2.
FISH labeling using BAC clones in sperm nuclei. (a) Sperm nucleus with a normal chromosome 5, displaying distinct green and red signals. (b) FISH results for BACs spanning the break points of the inverted chromosome 9 with one yellow and one collocated red/green spots. (c) Duplicated/deficient chromosome with one red or one combined red/green signal. (d) Dicentric chromosome with one red and one collocated red/green signal.
Data on meiotic segregation in PAI heterozygous are limited for male carriers. Only 10 male carriers of PAI were studied to date. As indicated in Table 2, large variations are observed in incidences of recombinant gametes (from 0 to 12.6%). The variations in the number of sperm counted (from 94 to 8158) and inverted segment size (from 5 to 41%) are also observed. A direct correlation between the size of inverted segments and the production of recombinants in sperm has been postulated.26, 27 This appears to be confirmed by our results obtained from large samples of segregation (>1000), as an increase of recombination rates is observed in parallel with the increase in inverted segment size (Table 2).
To date, two PAIs of chromosome 14 with almost similar break points and inverted segment size have been reported. Nevertheless, no recombinant was observed in the study of Martin,19 whereas we found 3.7% chromosome 14 recombinants. The difference in the sample size in both studies could explain the lack of recombinants in the study of Martin.19 Only 120 sperm metaphases were scored, whereas in the present study, 7670 sperm nuclei were analyzed. The variations in recombinants in small and large sample sizes were also observed by Jaarola et al.28 A significant presence of recombinants in the present study of PAI of chromosome 14 could be due to the higher rates of recombination in shorter chromosomes than in the larger chromosomes.29 In the case of chromosome 14, the recombination rate has been estimated as 1.36 cM/Mb. Sun et al30 found a mean of 2.03 sites of meiotic exchange in the q arms of chromosome 14, which is also higher than that in the q arms of other chromosomes.
Our detailed segregation analysis of three PAIs shows that each inversion displays a significant rate of unbalanced gametes even with an inverted segment size less than 50% (Table 2), contrary to the assumption drawn from previous sperm studies that the production of recombinant gametes requires a minimum segment proportion of 50%.26, 27 This needs to be confirmed on other inversions. However, this is consistent with the report of patients with recombinant chromosomes resulting from small inversions (<45 Mb and <25% of total length of chromosome).31, 32 The direct meaning of this observation in sperm could be that conventional sperm FISH assay using centromeric or telomeric DNA probes is definitively insufficient for a reliable segregation analysis of inversions because they can lead to underestimated rates of imbalances in sperm and then to erroneous genetic counseling of male inversion carriers.
Both chromosomes 5 and 14 are among the chromosomes most frequently implicated in PAI.1 The review of PAI studies has indicated that some particular chromosomal regions could be more prone to breakage and recombination. Ashley33 proposed that in inversions, both loop formation and the potential for recombination occur only when both break points are located in G-light chromosomal bands. Of the 10 PAIs analyzed in sperm to date (Table 2), a majority of them displayed break points in G-light domains. Manvelyan et al34 also observed the preferential appearance of the break points in the G-light domains in chromosomal rearrangements. On chromosome 14, breakages in G-light bands 14q13, 14q24 and 14q32 appear to be more frequent.1 Indeed, there is some evidence that certain particular genomic features might facilitate or predispose to chromosomal rearrangements such as translocations or inversions.35 The sequences around the rearrangement break points are frequently enriched by low copy repeats (LCRs) predisposing to nonallelic homologous recombination (NAHR), as reported by Giorda et al36 for the recurrent inv dup del(8p). Break points can be also colocalized with fragile sites, which promote genomic instability and double-strand breaks. The breakpoint 14q23.2 was found adjacent to the fragile site FRA14B. No LCRs or segmental duplications were found in the breakpoint regions of the PAI of chromosomes 5 and 14. This shows that for these two PAIs, LCRs were not involved in the formation of the inversion. Recently, Flores et al37 have identified in human chromosomes, several intrachromosomal identical reversed repeats of at least 400 nucleotides in length, which lead to chromosomal inversions by NAHR. These potential recombinogenic inverted sequences are distributed among all human chromosomes, suggesting that inversion rearrangements resulting from NAHR occur in a relatively high frequency in the human genome but preferentially on a nonvisible submicroscopic level.38, 39 These genomic rearrangements might play a prominent role in human genetic disease. Our understanding of all these structural genomic features as well as the growing number of discovered individual variations and their chromosomal consequences is still limited. The genomic rearrangements are potentially reversible and thus they do not permanently compromise on either the structure or the dynamics of the genome, but some of these genomic variations could be more deleterious because of position effect, direct disruption of critical genes or interaction with chromatid packaging. There is still more to learn about the genome structural features predisposing some chromosomes to rearrangements.
For PAI, the standard statement has it that there is a negligible risk of producing viable unbalanced offspring. However, reports of patients with recombinant chromosomes and the identification of various alternate mechanisms for recombinant generation (U-loop recombination, breakage and reunion of sister chromatids) refute the complete harmlessness of PAI. The formation of a dicentric chromosome during meiosis should not be considered as a rare or a benign event. Our detailed segregation analysis indicates that in male carriers, the production of imbalances may be significant. In oogenesis, Koelher et al40 showed that segregation attempts in dicentrics could result in the premature loss of sister centromere cohesion and the segregation of an intact dicentric. Such meiotic behavior could explain the cases of viable stable dicentrics of maternal origin reported to date.3, 4, 5, 6 Consequently, the production and the transmission of recombinant chromosomes through meiosis must be kept in mind when counseling carriers of PAI. In addition, efforts must be made to accurately identify all possible recombinants and distinguish between PAI and intrachromosomal insertion, which displays a significantly higher risk of genetic imbalance (from 15 to 30%) than PAI.13 The in situ mapping of chromosomal break points by probe contigs, as presented herein, constitutes the first and efficient approach to avoid erroneous diagnosis. Combined with high-resolution genomic screening, this approach may yield new insights into the mechanisms involved in the generation of unbalanced rearrangements.
Acknowledgments
Supported in part by the IZKF Jena, DFG (436 ARM 17/11/06, LI 820/15-1), DAAD (A0704616/Ref326), Ferring Pharmaceuticals and Organon France. We thank the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply and verification.
References
- Madan K. Paracentric inversions: a review. Hum Genet. 1995;96:503–515. doi: 10.1007/BF00197403. [DOI] [PubMed] [Google Scholar]
- Fryns J, Kleczkowska A, kubien E, van den Berghe H.Structural chromosome rearrangements in couples with recurrent fetal wastage: the leuven experiencein Sandberg A (ed): The Cytogenetics of Mammalian Autosomal Rearrangements New York: Alan R Liss; 1988. Vol 8, pp453–470. [Google Scholar]
- Worsham MJ, Miller DA, Devries JM, et al. A dicentric recombinant 9 derived from a paracentric inversion: phenotype, cytogenetics, and molecular analysis of centromeres. Am J Hum Genet. 1989;44:115–123. [PMC free article] [PubMed] [Google Scholar]
- Mules EH, Stamberg J. Reproductive outcomes of paracentric inversion carriers; report of a liveborn dicentric recombinant and literature review. Hum Genet. 1984;67:126–131. doi: 10.1007/BF00272986. [DOI] [PubMed] [Google Scholar]
- Whiteford ML, Baird C, Kinmond S, Donaldson B, Davidson HR. A child with bisatellited, dicentric chromosome 15 arising from a maternal paracentric inversion of chromosome 15q. J Med Genet. 2000;37:E11. doi: 10.1136/jmg.37.8.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort G, Blanchet P, Belgrade N, et al. Stable dicentric duplication-deficiency chromosome 14 resulting from crossing-over within a maternal paracentric inversion. Am J Hum Genet. 2002;113:333–338. doi: 10.1002/ajmg.b.10720. [DOI] [PubMed] [Google Scholar]
- Phelan MC, Stevenson RE, Anderson EV., Jr Recombinant chromosome 9 possibly derived from breakage and reunion of sister chromatids within a paracentric inversion loop. Am J Med Genet. 1993;46:304–308. doi: 10.1002/ajmg.1320460313. [DOI] [PubMed] [Google Scholar]
- Yang SP, Bidichandani SI, Figuera LE, et al. Molecular analysis of deletion (17)(p11.2p11.2) in a family segregating a 17p paracentric inversion: implications for carriers of paracentric inversions. Am J Hum Genet. 1997;60:1184–1193. [PMC free article] [PubMed] [Google Scholar]
- Feldman GL, Weiss L, Phelan M, Shroer RJ, Van Dyke DL. Inverted duplication of 8p: ten new patients and review of the literature. Am J Med Genet. 1993;47:482–486. doi: 10.1002/ajmg.1320470410. [DOI] [PubMed] [Google Scholar]
- Mitchell JJ, Vekemans M, Luscombe S, et al. U-type exchange in a paracentric inversion as a possible mechanism of origin of an inverted tandem duplication of chromosome 8. Am J Med Genet. 1994;49:384–387. doi: 10.1002/ajmg.1320490406. [DOI] [PubMed] [Google Scholar]
- Pettenati MJ, Rao PN, Phelan MC, et al. Paracentric inversions in humans: a review of 446 paracentric inversions with presentation of 120 new cases. Am J Med Genet. 1995;55:171–187. doi: 10.1002/ajmg.1320550207. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Callen DF, Gardner RJM. Paracentric inversions do not normally generate monocentric recombinant chromosomes. Am J Med Genet. 1995;59:390. doi: 10.1002/ajmg.1320590323. [DOI] [PubMed] [Google Scholar]
- Madan K, Nieuwint AW. Reproductive risks for paracentric inversion heterozygotes: inversion or insertion? That is the question. Am J Med Genet. 2002;107:340–343. doi: 10.1002/ajmg.10173. [DOI] [PubMed] [Google Scholar]
- Cheng EY, Chen YJ, Disteche CM, Gartler SM. Analysis of a paracentric inversion in human oocytes: nonhomologous pairing in pachytene. Hum Genet. 1999;105:191–196. doi: 10.1007/s004399900120. [DOI] [PubMed] [Google Scholar]
- Brown GM, Leversha M, Hulten M, Ferguson-Smith MA, Affara NA, Furlong RA. Genetic analysis of meiotic recombination in humans by use of sperm typing: reduced recombination within a heterozygous paracentric inversion of chromosome 9q32–q34.3. Am J Hum Genet. 1998;62:1484–1492. doi: 10.1086/301863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F, Andréo B, Puechberty J, Lefort G, Sarda P.Analysis of sperm aneuploidy by PRINSin Pellestor F (ed): PRINS and in situ PCR protocolssecond edn.Totowa: Humana Press; 200649–60. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Moradkhani K, Mrasek K, et al. Breakpoint characterization: a new approach for segregation analysis of paracentric inversion in human sperm. Mol Hum Reprod. 2007;13:751–756. doi: 10.1093/molehr/gam048. [DOI] [PubMed] [Google Scholar]
- Martin RH. Sperm chromosome analysis in a man heterozygous for a paracentric inversion of chromosome 7 (q11q22) Hum Genet. 1986;73:97–100. doi: 10.1007/BF00291594. [DOI] [PubMed] [Google Scholar]
- Martin RH. Sperm chromosome analysis in a man heterozygous for a paracentric inversion of chromosome 14 (q24.1q32.1) Am J Hum Genet. 1999;64:1480–1484. doi: 10.1086/302363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine DH, Whitman-Elia G, Best RG, Edwards JG. Paternal paracentric inversion of chromosome 2: a possible association with recurrent pregnancy loss and infertility. J Assist Reprod Genet. 2000;17:293–296. doi: 10.1023/A:1009466501616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton E, Vidal F, Egozcue J, Blanco J. Genetic reproductive risk in inversion carriers. Fertil Steril. 2006;85:661–666. doi: 10.1016/j.fertnstert.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Vialard F, Delanete A, Clement P, Simon-Bouy B, Aubriot FX, Selva J. Sperm chromosome analysis in two cases of paracentric inversion. Fertil Steril. 2007;87:418. e1–418. e5. doi: 10.1016/j.fertnstert.2006.05.087. [DOI] [PubMed] [Google Scholar]
- Anton E, Blanco J, Egozcue J, Vidal F. Risk assessment and segregation analysis in a pericentric inversion inv6p23q25 carrier using FISH on decondensed sperm nuclei. Cytogenet Genome Res. 2002;97:149–154. doi: 10.1159/000066603. [DOI] [PubMed] [Google Scholar]
- Mikhaail-Philips MM, McGillivray BC, Hamilton SJ, et al. Unusual segregation products in sperm from a pericentric inversion 17 heterozygote. Hum Genet. 2005;117:357–365. doi: 10.1007/s00439-004-1245-0. [DOI] [PubMed] [Google Scholar]
- Chantot-Bastaraud S, Ravel C, Berthaut I, et al. Sperm-FISH analysis in a pericentric chromosome 1 inversion, 46, XY, inv (1) (p22q42), associated with infertility. Mol Hum Reprod. 2007;13:55–59. doi: 10.1093/molehr/gal094. [DOI] [PubMed] [Google Scholar]
- Anton E, Blanco J, Egozcue J, Vidal F. Sperm studies in heterozygote inversion carriers: a review. Cytogenet Genome Res. 2005;111:297–304. doi: 10.1159/000086903. [DOI] [PubMed] [Google Scholar]
- Morel F, Laudier B, Guerif F, et al. Meiotic segregation analysis in spermatozoa of pericentric inversion carriers using fluorescence in situ hybridization. Hum Reprod. 2007;22:136–141. doi: 10.1093/humrep/del317. [DOI] [PubMed] [Google Scholar]
- Jaarola M, Martin Rh, Ashley T. Direct evidence for suppression of recombination within two pericentric inversions in humans: a new sperm FISH technique. Am J Hum Genet. 1998;63:218–224. doi: 10.1086/301900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Sun F, Oliver-Bonet M, Liehr T, et al. Human male recombination maps for individual chromosomes. Am J Hum Genet. 2004;74:521–531. doi: 10.1086/382138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone R, Mattina T, Giorda R, et al. Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet. 2006;43:e19. doi: 10.1136/jmg.2005.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Swensen JJ, Maxwell T, Rope A, Brothman AR, Chen Z. A new genomic mechanism leading to cri-du-chat syndrome. Am J Med Genet A. 2006;140:2714–2720. doi: 10.1002/ajmg.a.31496. [DOI] [PubMed] [Google Scholar]
- Ashley T. G-band position effects on meiotic synapsis and crossing over. Genetics. 1988;118:307–317. doi: 10.1093/genetics/118.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manvelyan M, Shreyer I, Hols-Herpertz I, et al. Forty-eight cases with infertility due to balanced chromosomal rearrangements: detailed molecular cytogenetic analysis of the 90 involved breakpoints. Int J Mol Med. 2006;19:855–864. doi: 10.3892/ijmm.19.6.855. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev. 2002;12:312–319. doi: 10.1016/s0959-437x(02)00304-0. [DOI] [PubMed] [Google Scholar]
- Giorda R, Ciccone R, Gimelli G, et al. Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat. 2007;28:459–468. doi: 10.1002/humu.20465. [DOI] [PubMed] [Google Scholar]
- Flores M, Morales L, Gonzaga-Jauregui C, et al. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- http://projects.tcag.ca/variation/ .
- Koehler KE, Millie EA, Cherry JP, et al. Sex-specific differences in meiotic chromosome segregation revealed by dicentric bridge resolution in mice. Genetics. 2002;162:1367–1379. doi: 10.1093/genetics/162.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]