Abstract
Lysinuric protein intolerance (LPI) is a rare autosomal inherited disease caused by defective cationic aminoacid transport 4F2hc/y+LAT-1 at the basolateral membrane of epithelial cells in the intestine and kidney. LPI is a multisystemic disease with a variety of clinical symptoms such as hepatosplenomegaly, osteoporosis, hypotonia, developmental delay, pulmonary insufficiency or end-stage renal disease. The SLC7A7 gene, which encodes the y+LAT-1 protein, is mutated in LPI patients. Mutation analysis of the promoter localized in intron 1 and all exons of the SLC7A7 gene was performed in 11 patients from 9 unrelated LPI families. Point mutation screening was performed by exon direct sequencing and a new multiplex ligation probe amplification (MLPA) assay was set up for large rearrangement analysis. Eleven SLC7A7-specific mutations were identified, seven of them were novel: p.L124P, p.C425R, p.R468X, p.Y274fsX21, c.625+1G>C, DelE4-E11 and DelE6-E11. The novel large deletions originated by the recombination of Alu repeats at introns 3 and 5, respectively, with the same AluY sequence localized at the SLC7A7 3′ region. The novel MLPA assay is robust and valuable for LPI molecular diagnosis. Our results suggest that genomic rearrangements of SLC7A7 play a more important role in LPI than has been reported, increasing the detection rate from 5.1 to 21.4%. Moreover, the 3′ region AluY repeat could be a recombination hot spot as it is involved in 38% of all SLC7A7 rearranged chromosomes described so far.
Keywords: lysinuric protein intolerance, SLC7A7 mutations, large deletions, AluY-mediated recombination, hot spot
Introduction
Lysinuric protein intolerance (LPI, MIM 222700) is a primary inherited aminoaciduria caused by defective dibasic amino acid transport activity, which affects renal reabsorption of cationic amino acids and its transport function in the small intestine. Lung, liver, immune system, muscle and brain are also affected to a varying extent in LPI patients.1 LPI has an autosomal recessive mode of inheritance and is relatively common in Finland, with a prevalence of 1/60 000. It also occurs in Southern Italy and Japan (prevalence of 1/50 000 in the northern part of Iwate).2, 3, 4 Sporadic cases have been described worldwide in other 22 countries.5, 6
Lysinuric protein intolerance is a multiorgan disease with a variety of clinical symptoms. Patients with LPI are usually asymptomatic while being breast fed, and symptoms appear only after weaning. After infancy, they reject a high-protein diet and show bone growth delay, osteoporosis, hepatosplenomegaly and muscle hypotonia. Most patients show normal mental development, but some have moderate retardation. About two-thirds of the patients have interstitial changes in chest radiographs, sometimes with acute or chronic respiratory insufficiency that can lead to fatal pulmonary alveolar proteinosis and multiple-organ dysfunction syndrome.1, 7 Recently, some patients with end-stage renal disease have been reported.8 Further symptoms, such as glomerulonephritis and erythroblastophagia, suggest that the immune system is affected in some patients.9
Lysinuric protein intolerance is diagnosed by urine hyperexcretion of dibasic amino acids, especially lysine, and by their poor intestinal absorption. As a result, plasma levels of dibasic amino acids are low in patients.1, 7, 9, 10 Arginine and ornithine are intermediates of the urea cycle that provide the carbon skeleton. Their reduced availability is responsible for hyperammonaemia, which is thought to cause acute symptoms such as nausea and vomiting. Protein malnutrition and lysine deficiency contribute to the patient's failure to thrive.9 Patients are treated with low-protein diet and supplementation of -citrulline, a urea cycle intermediate, absorbed by neutral amino acid transporters, which is used to correct their urea cycle functional deficiency. Low-dose oral lysine supplementation is potentially beneficial to patients with LPI starting at an early age.11
The human gene responsible for LPI, SLC7A7 (MIM 603593; RefSeq: NM_003982), was localized to 14q11.2 by linkage.12, 13, 14, 15 It contains 11 exons (9 coding exons), 10 introns and two promoter regions spanning ∼46.5 kb of the genomic DNA.3, 4, 16 The cDNA is 2477 bp long with a 527 bp 5′-UTR and a 385 bp 3′-UTR, with a polyA site located at 2447.6 It encodes the y+LAT1 protein (RefSeq: NP_003973), which has 511 predicted amino acids and a molecular weight of ∼56 kDa.17 y+LAT1 heterodimerizes with 4F2hc to express system y+L amino acid transport activity in the basolateral plasma membrane of the epithelial cells of the renal proximal tubule and the small intestine.17 System y+L mediates the electroneutral exchange of dibasic amino acids (efflux) for neutral amino acids and sodium (influx).17
Since now a total of 44 different SLC7A7-specific mutations have been identified in 131 patients including all different types of mutations distributed along the entire gene.5, 6 Five polymorphisms within the SLC7A7 coding region and two in the 5′-UTR have also been identified.6 A founder effect mutation has been demonstrated only in Finland, where LPI patients share the same homozygous mutation, c.895-2A>T.2, 14 LPI patients show extreme variability in clinical presentation, and no genotype–phenotype correlations have been defined.18 This phenotypic variability and the lack of a specific clinical presentation have led to various misdiagnoses.6
Here we describe the spectrum of mutations in the SLC7A7 gene in nine unrelated LPI families. An SLC7A7 gene-specific multiplex ligation probe amplification (MLPA) assay was set up for the first time to search for large rearrangements in SLC7A7 gene. Seven novel mutations have been found: two missense, a nonsense, a splice donor, a nucleotide duplication and two large deletions originated by AluY-mediated homologous recombination.
Materials and methods
Patients
We studied 11 LPI patients, and their relatives when available, from nine unrelated families with different geographic origins: Spanish (three), Chilean (two, half brothers), Moroccan (two), English-Argentinean (one), French (one) and Greek (two, brothers). According to the referring centres, the main clinical features of these patients were compatible with those of LPI (Table 1). Informed consent was obtained from all patients, their parents and 50 healthy controls. This research was prospectively reviewed and approved by the Ethics Committee of the IDIBELL.
Table 1. Clinical features of LPI patients.
| Patient no. | 1 | 2 | 3 | 4 | 5 | 6 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Age | 6 years | 11 years | 2 years | 14 years | 12 years | 13 years | 9 months | 14 months | 15 years |
| Sex | M | F | M | M | F | F | F | F | M |
| Basic hyperaminoaciduria | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Basic hypoaminoacidaemia | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Developmental delay | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Protein intake aversion | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Hepatosplenomegaly | Yes (mild) | Yes (mild) | Yes (mild) | Yes | Yes | Yes | No | No | Yes (mild) |
| Osteoporosis | Yes | Yes | — | Yes | Yes | — | — | No | Yes |
| Hypotony | No | No | Yes | No | No | Yes | Yes | Yes | — |
| Anaemia | Yes | No | Yes | Yes | Yes | — | No | No | — |
| Mental retardation | — | No | — | — | Yes | Yes | — | No | — |
| Pulmonary insufficiency | No | No | — | — | No | Yes | — | — | — |
Genomic DNA amplification, sequencing and mutation analysis
DNA was extracted from peripheral leukocytes from whole blood using standard protocols. Mutational screening of the whole coding region and all intron/exon boundaries was performed by PCR and direct sequencing, with PCR conditions and primers reported earlier.3 The promoter localized in intron 1, which is the active promoter in tissues with strong defects in LPI patients,16 was also amplified with primers (5′-CTGGCCTGATTTCCTCATATT-3′) and [5′-GAGGGTTAGCAAGGTAAGTGG-3′), following standard methods. The reaction started with an initial denaturation of 5 min at 94°C, followed by 36 cycles of 94°C 30 s, 58°C 30 s, 74°C 1 min and 5 min at 74°C.
All newly described mutations were checked on 100 control chromosomes to discard polymorphisms. All new mutations follow the nomenclature of the HGVS (http://www.hgvs.org/mutnomen/) and have been introduced in the HGMDB database (http://www.hgmd.org/).
MLPA analysis
Probes within the MLPA set were designed following instructions by MRC Holland (http://www.mlpa.com/pages/support_desing_synthetic_probespag.html). The presence of repetitive sequences was excluded using the BLAT program from UCSC website (http://genome.ucsc.edu),19 and regions with SNPs or known LPI-specific mutations were avoided (http://www.ensembl.org and reported LPI data) (Supplementary Table 1). Oligonucleotides were ordered from Sigma Genosys (UK, http://www.sigma-genosys.com). MLPA reactions were performed essentially as described20 with hybridization and ligation reagents purchased from MRC Holland. Genomic DNA (150–200 ng) in a final volume of 5 μl was heated at 98°C for 5 min. After cooling to room temperature, 1.5 μl probe mix and 1.5 μl SALSA hybridization buffer were added to each sample, heat-denatured at 95°C for 1 min, followed by hybridization for 16 h at 60°C. Ligation was performed at 54°C by adding 32 μl ligation mix. After 15 min, the reaction was stopped by heat inactivation at 95°C for 5 min. Amplifications were carried out in a 25 μl final volume containing 5 μl of ligation product, 2 μl of SALSA PCR buffer (MRC Holland, Amsterdam), 1 μl of Taq dilution buffer (Roche Diagnostics), 180 n of each MLPA primer (Sigma), 1 m of dNTPs (Ecogen, Spain) and 1.25 U of Taq Polymerase (Roche Diagnostics). The products were analysed on an ABI PRISM 3100 genetic analyser according to the manufacturers' instructions. MLPA data were analysed by visual examination of the peak profiles and by using the relative peak height (RPH) method as previously described.21 Heterozygous deletions and duplications should show an RPH of ∼0.5 and 1.5, respectively, and no peak reveals a homozygous deletion.
Characterization of large rearrangements
Eleven PCR primers were designed along nonrepetitive sequences from intron 3, intron 5 and the 3′ region, obtained in the RepeatMasker Web Server (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) (primers sequences are available upon request). PCR on patient nos. 3 and 5, with all possible combinations of forward primers from the respective intron and reverse primers from the 3′ region, was performed. Positive amplification was obtained with primers: LPI_I3F7 (5′-CTGAGCATCTGATCCAGAGT-3′) and LPI_3′R1 (5′-CAGTCCTAGGAACTGTGGCA-3′) for DelE4-E11, and E5F (5′-TGTCAGTCCTTTACTTGACCATC-3′) and LPI_3′R1 for DelE6-E11 characterization. PCR was performed as described above. An initial denaturation of 5 min at 94°C was followed by 10 cycles at 94°C 20 s, 58°C 20 s, 72°C 15 min and 30 cycles at 90°C 20 s, 58°C 20 s, 72°C 15 min and a final extension of 7 min at 72°C.
Results
Clinical data
All patients had plasma concentrations and amino acid urinary excretion patterns characteristic of LPI. Some patients had elevated serum ferritin and lactate dehydrogenase concentrations. They had shown hyperammonaemia after high-protein meals, and most of them had hepatosplenomegaly, developmental delay, bone growth delay, osteoporosis and muscle hypotonia (Table 1). Some patients presented anaemia, pulmonary insufficiency and/or mental retardation. All patients are under treatment with -citrulline (treatment of patient no. 2 has been already reported22).
Mutation analysis of SLC7A7
Eleven SLC7A7 mutations were found, seven of which are novel.16 The mutations comprise two frameshift (c.820dupT, c.1185_1188delTTCT), three nonsense (p.R410X, p.Y457X, p.R468X), three missense (p.L124P, p.L334R, p.C425R), a splice donor (c.625+1G>C) and two large rearrangements identified by MLPA (DelE4-E11, DelE6-E11). All mutations segregated with the disease status in the respective families (Figure 1c). The novel mutations are p.L124P, p.C425R, p.R468X, c.625+1G>C, c.820dupT, and the two large deletions are DelE4-E11 and DelE6-E11 (Figure 1 and Table 2). None of them were detected in 100 control chromosomes tested (data not shown). In addition, none of the SLC7A7 sequence changes underlying these novel mutations are listed in the NCBI Single Nucleotide Polymorphism database (dbSNPs; http://www.ncbi.nlm.nih.gov/projects/SNP) or in Ensembl (http://www.ensembl.org). Seven silent polymorphisms were also found, some of them reported earlier2, 4 (Supplementary Table 2).
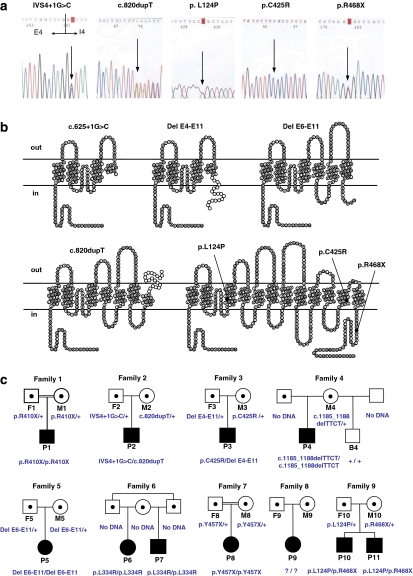
Figure 1.
New SLC7A7 point mutations in patients with LPI and segregation analysis of all the mutations detected in LPI families. (a) Electropherogram sections showing the new point mutations: p.L124P, p.C425R, p.R468X, c.625+1G>C and c.820dupT. (b) Scheme of the predicted mutant proteins in the topology model of human y+LAT-1 of 12 transmembrane domains. All the novel mutations but two encode truncated proteins, if translated. Interestingly, c.625+1G>C and DelE4-E11 mutations would encode nearly the same protein lacking the last nine transmembrane domains, but the differences at the mRNA level (presence or absence of polyA signal, 3′-UTR and so on) could give differences in its stability and translation. Notice that two of the frameshift mutations, Del E4-E11 and c.820dupT, have addition of missense residues (white circles). (c) Segregation of all the mutations found in 11 patients with LPI by direct sequencing. Parents of patient nos. 1 and 7 are closely consanguineous and patient nos. 6 and 7 have the same mother and their fathers are brothers. In families 4 and 6, the segregation could not be performed because there was no DNA available from the relatives, but normal MLPA pattern of these patients (Supplementary Table 1) confirmed the homozygosity of these patients. Square symbols are men and circles are women. The filled symbols are affected individuals and the half-filled symbols are obligated carriers. B, brother; F, father; M, mother; P, patient; +, wild-type allele.
Table 2. Mutations in the SLC7A7 gene causing LPI in the patients analysed.
| Patient no. | Nucleotide change | Predicted protein change | Status | Exon/Intron | Mutation type | Geographic origin |
|---|---|---|---|---|---|---|
| 1 | c.1228C>T | p.R410X | Homozygote | 9 | Nonsense | Moroccan |
| 2 | c.820dupT c.625+1G>C | p.Y274fsX21 p.C167_G209delinsX | Compound heterozygote | 6 4 | Frameshift Splice site | English/Argentinean |
| 3 | c.1273T>C c.500−4294_1908+1028 del12136 | p.C425R p.L168_N511delfsX19 | Compound heterozygote | 10 I3-3′region | Missense Large rearrangement | Spanish |
| 4 | c.1185_1188delTTCT | p.S396fsX121 | Homozygote | 9 | Frameshift | Spanish |
| 5 | c.771−848_1908+718 del4647 | p.N258_N511delinsX | Homozygote | I5-3′region | Large rearrangement | Spanish |
| 6 and 7 | c.1001T>G | p.L334R | Homozygote | 8 | Missense | Chilean |
| 8 | c.1371C>A | p.Y457X | Homozygote | 10 | Nonsense | Moroccan |
| 10 and 11 | c.371T>C c.1402C>T | p.L124P p.R468X | Compound heterozygote | 3 10 | Missense Nonsense | Greek |
Figure 1 and Table 2 show the novel mutations found in this group of patients. c.625+1G>C mutation is a G-to-C transversion that modifies the splice GT donor site of intron 4. Interestingly, the mutation is located in the same nucleotide position as c.625+1G>A reported earlier, which was demonstrated to cause exon 4 skipping,4 suggesting aberrant splicing for this mutation. This would result in a truncated y+LAT1 protein with only three transmembrane domains and the re-entrant loop between TMD II and III (p.C167_G209del42insX). The c.820dupT mutation in exon 6 creates a frameshift after residue I273 with the addition of 20 missense residues before the first stop codon (p.Y274fsX21) eliminating half of the protein, resulting in a truncated protein with six TMD. The p.C425R mutation is caused by a C-to-T transition at nucleotide position 1228 of the cDNA in exon 10, resulting in a nonconservative amino acid exchange from a slightly polar amino acid (cystine) to a positively charged amino acid (arginine) at position 425. The substituted cysteine residue is located in the transmembrane domain XI (residues 419–438) of y+LAT-1, introducing a positively charged residue. The p.L124P mutation is caused by a T-to-C transition at nucleotide position 371 of the cDNA in exon 3, which is located in the transmembrane domain III (residues 117–140) of y+LAT-1. The p.R468X is a C-to-T transition at nucleotide position 1402 of the cDNA in exon 10. The premature stop codon should originate a truncated protein lacking the last 44 cytoplasmatic residues, if transcribed.
MLPA analysis
The presence of SLC7A7 large rearrangements was assessed by MLPA with specific probes to find out copy number changes in these patients. Figure 2 shows the abnormal MLPA patterns found in two LPI families. In family no. 3, the patient and his father bore a deletion of exons 4–11, whereas the mother, who bore p.C425R mutation in heterozygosis, showed a normal MLPA pattern. In family no. 5, both parents showed half-dosage for exons 6–11, whereas the patient showed no peak for these exons. The rest of the patients and relatives and 10 control individuals showed a normal MLPA pattern (Supplementary Table 3).
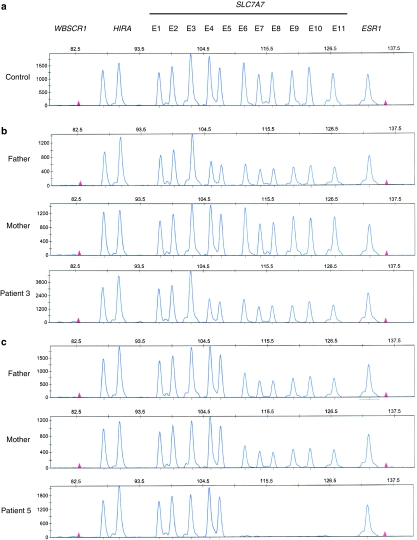
Figure 2.
MLPA patterns of families 3 and 5. Samples were subjected to MLPA with 11 specific oligonucleotide probes for SLC7A7 exon sequences. Three control probes for genes located on different chromosomes were also added in the assay as external controls. Each individual region's peak height is normalized by dividing it by the sum of control gene peak heights (WBSCR1, HIRA, ESR1). Each normalized peak height is then divided by the corresponding average normalized peak height from control samples to obtain the RPH. (a) Normal SLC7A7 MLPA pattern obtained from a control individual. (b) Deletion of the two-thirds of the SLC7A7 gene detected by MLPA in family no. 3. The father and the patient show an abnormal pattern. MLPA peak pattern from the mother is the same as that of the control. (c) Deletion of the last six exons of SLC7A7 gene detected by MLPA in family no. 5. Both parents carry the deletion in heterozygosis and the patient is a homozygote.
Breakpoint characterization of the novel large rearrangements
Breakpoints of the two large SLC7A7 deletions were mapped by standard PCR across the junction fragments with primers that specifically amplify the mutated allele in both cases. In patient no. 3, a 1 kb fragment was amplified, purified and sequenced using primers LPI-I3F3 and LPI-3′R1, showing a deletion of 12 136 bp with a breakpoint at 4294 bp upstream of exon 4 and 1028 bp downstream of exon 11 (c.500−4294_1908+1028del12136) (Figure 3a). The protein product expected from the 12 kb deletion, if translated, would have a frameshift after residue C167 with the addition of 18 missense residues before the first stop codon (p.L168_N511delfsX19), lacking transmembrane domains IV–XII (Figure 1b). In patient no. 5, a 3 kb fragment was amplified with primers LPI-23FG and LPI-3′R1 showing a deletion of 4647 bp with a breakpoint at 848 bp upstream of exon 6 and 718 bp downstream of exon 11 (c.771−848_1908+718del4647) (Figure 3b). The protein product of the 4.6 kb deletion would have a stop codon after residue R257 (p.N258_N511delinsX), lacking transmembrane domains VII–XII (Figure 1b).
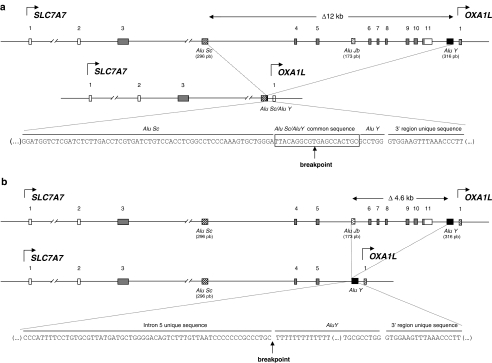
Figure 3.
Scheme of the two large deletions in SLC7A7 originated by the crossing over between two direct Alu repeats, found in LPI patient nos. 3 and 5. (a) Genomic scheme of the SLC7A7 gene and the 12 kb deletion (DelE4-E11) found in patient no. 3, with a breakpoint at 4294 bp upstream exon 4 and 1028 bp downstream exon 11 (c.500−4294_1908+1028del12136). Notice that the breakpoint could be generated in any of the nucleotides in the box, as this 20 nt sequence is the same in both the Alu repeats involved in this genomic rearrangement. The mutation nomenclature localizes the breakpoint at the 3′ end of the 20 nt sequence. (b) The 4.6 kb deletion (DelE6-E11) found in patient no. 5, had the breakpoint at 848 bp upstream of exon 6 and 718 bp downstream of exon 11 (c.771−848_1908+718del4647). In DelE4-E11, the breakpoint was at the 3′ end of AluY, whereas in DelE6-E11, the breakpoint was located at the first nucleotide of the AluY sequence.
Both large deletions were originated by the crossing over between two direct Alu repeats: DelE4-E11 was originated by recombination of a 296 bp AluSc in intron 3 and a 316 bp AluY in the SLC7A7 3′ region, whereas DelE6-E11 was originated by recombination of a 173 bp AluJb in intron 5 and the same AluY repeat in the SLC7A7 3′ region (Figure 3). BLAST two sequences of AluSc-AluY and AluJb-AluY revealed 86 and 80% nucleotide identity, respectively. Analysis of the genomic regions implicated in these deletions showed a high content of repetitive elements. Intron 3 of SLC7A7 gene contains 70 complex repetitive elements (47 Alu, 4 MIRs, 12 LINEs, 4 LTR elements, 2 MERs), which represent 67% of the 32.8 kb of the sequence. Intron 5 has six complex repetitive elements (four Alu and two MIRs) along 2.4 kb, which represent 45% of the intron, whereas in the SLC7A7 3′ region, there are eight repetitive sequences (five Alu and three LINEs) along the 5 kb downstream exon 11.
Discussion
To date, 51 LPI-specific SLC7A7 mutations have been described (this study and Cimbalistiene et al5 and Sperandeo et al6), including 21 missense, 6 nonsense, 5 splice site, 4 small duplications/insertions, 10 small deletions and 5 large genomic rearrangements. The mutation screening of SLC7A7 gene performed in this study revealed seven novel mutations and four mutations described earlier in eight LPI-unrelated families, which explain 89% of the studied alleles. Patient no. 9 bore only three polymorphisms in heterozygosis: rs8019656:G>A, c.1-86C>T and c.159A>G. She showed the mildest phenotype with good clinical evolution, indicating that she could have been misdiagnosed. Alternatively, missing mutations in this patient might be located in unexplored intronic regions. We considered the possibility of finding mutations in the heavy-chain 4F2hc very unlikely, as no LPI-associated mutation had been found in SLC3A2,18 and Slc3a2 knockout mouse is lethal.23 Indeed, 4F2hc serves, in addition to system y+L, as the heavy subunit of five other amino acid transporters and has a role in β1-integrin function.24
All the new mutations identified but two encode for severely truncated proteins, if translated, suggesting a dramatic loss of transport function in these cases. The novel large deletions (DelE6-E11 and DelE4-E11) were associated with the most severe phenotypes. A plausible explanation would be that truncated transcripts lacking 3′-UTR sequence and polyA signal would end up in rapid degradation.25 In contrast, frameshift (c.820dupT), splice-site (c.625+1G>C) and nonsense (p.R468X) mutations associate with less severe phenotypes in this group of patients. These mutations also introduce premature termination codons into the mRNA, which in most cases result in degradation through a mechanism known as nonsense-mediated decay and loss of function of the mutated allele.26 However, c.1185_1188delTTCT mutation found in homozygosis and c.820dupT mutation found in heterozygosis are associated with a milder phenotype, suggesting that some of the truncated transporter may reach the plasma membrane and may retain partial activity in these patients. The missense mutation p.L124P is located in the transmembrane domain III in a highly conserved amino acid region among the known eukaryotic members of LAT family. The missense mutation, p.C425R, introduces a charged residue in transmembrane domain XI. None of the >80 known eukaryotic members of the LAT family, to which y+LAT1 belongs, has a charged residue in this position or in the entire transmembrane domain XI.27 This suggests a loss of transport function for these mutated transporters.
A detection system for SLC7A7 rearrangements by MLPA was established, and two novel large deletions of 12 and 4.6 kb were detected in two families. Interestingly, both deletions were originated by Alu-mediated recombination with identical AluY repeat at the 3′ region. These deletions involve the 5′ region of the adjacent gene OXA1L, as AluY element localizes at 388 bp of this gene, overlapping the SLC7A7 3′ region. OXA1L gene encodes a mitochondrial integral membrane protein required for the correct biogenesis of F(1)F(o)-ATP synthase and NADH:ubiquinone oxidoreductase.28 Considering that the homozygote patient with DelE6-E11 (patient no. 5) has a severe but characteristic LPI phenotype, the deletion of this sequence from the OXA1L 5′ region may not affect the transcription of this contiguous gene, but further enzymatic deficiency studies of the OXPHOS system should be performed in this patient to confirm this.
Alu repeats are short interspersed nuclear elements, which are the most common repetitive DNA units dispersed throughout the genome. The 280-bp repeat unit occurs approximately once every 4 kb in the human genome.29 Regions with high Alu repeat content are prone to recurrent genomic rearrangements through Alu–Alu recombination events.30 In fact, there are several inherited disorders for which Alu-mediated recombination is a common cause.31 Given the high Alu density in introns 3, 5 and the 3′ region, and the high degree of homology between Alu repeats, a recombination event could potentially occur anywhere within this region. However, the same AluY at the 3′ region is involved in 38% (three out of eight) of all unrelated chromosomes with SLC7A7 large rearrangements described so far,6 suggesting that it could be a recombination hot spot, as it has been observed at the duplication and deletion breakpoints in a number of human genetic diseases.30, 31, 32, 33 The Alu core sequence itself has been suggested to promote DNA-strand exchange and genomic rearrangement, and it has striking sequence similarity to chi (which has been shown to stimulate recBCD-mediated recombination in Escherichia coli).30 Although repeat DNA sequences may predispose to abnormal chromosome pairing and unequal crossing over, the fact that this region could be a hot spot implies that there must be other cellular mechanisms (sequence signals close to this AluY, chromatin structure33 and so on) that may play a more prominent role in the initiation of chromosomal rearrangements.
Our results demonstrate that the percentage of chromosomes with large rearrangements in SLC7A7 gene in this study is more than four times higher (21.4%) than that the previously reported by all other non-Finnish groups of patients (5.1%).5, 6 This demonstrates that deletions within this gene are more common than those have been reported so far, suggesting that the latter percentage is probably underestimated. In fact, there are some LPI patients with no mutated alleles reported in the literature: a patient with suspected LPI in which no mutations were found2 and five patients with only one mutant allele identified.6 In these cases, the screening strategy was based on sequencing of coding regions and intronic boundaries. These patients could have large rearrangements considering our results, although mutations in the intronic regions cannot be discarded. In addition, some of the previously reported homozygotes with no segregation performed could in fact be compound heterozygotes with a large deletion in the other allele. The MLPA technique has allowed us not only to find novel large rearrangements but also to discard hemizygosis in families in which there was no DNA from the relatives.
In conclusion, our study shows that deletions of the SLC7A7 gene are a common cause of LPI in non-Finnish populations, and therefore we highly recommend this MLPA assay for molecular diagnosis of LPI patients, as it covers the whole gene in one simple and reproducible assay. We estimate that it would detect around 15–20% partial or whole gene deletions in non-Finnish patients. Our data also suggest that recombination between Alu repeat elements is likely to be the major mechanism underlying SLC7A7 gene rearrangements. Moreover, the AluY repeat localized at 3′ of the SLC7A7 gene is involved in the majority of large rearrangements described in LPI patients.
Acknowledgments
We thank Helena Kruyer for editorial help. The samples and clinical information were kindly provided by Dr Lambruschini, Hospital General de Granollers, Spain; Dr Gómez and Dr Vilaseca, Hospital Sant Joan de Déu de Barcelona, Spain; Dr Gutiérrez Junquera and Dr Vilaseca, Complejo Hospitalario Universitario de Albacete, Spain; Dr Peña Quintana, Hospital Universitario Materno-Infantil de Canarias, Spain; Dr Antonio Baldellou, Hospital Infantil Miguel Servet de Zaragoza, Spain; Dr Quitral Cuevas, Valparaíso, Chile; Dr Vianey-Saban, Hôpital de Lyon, France; Drs Michelakakis and Xaidara, Institute of Child Health and University of Athens, Greece. This work was supported by the MEC (BFU2006-14600-C02-01/02/BMC and SAF2003-08940-01/02), EU (EUGINDAT; LSHM-CT-2003-502852) and the Generalitat de Catalunya (2006 SGR00018 and 2005 SGR00947).
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Simell O.Lysinuric protein intolerance and other cationic aminoaciduriasin Scriver CR, Beaudet AL, Sly WS, Valle D (eds):The Metabolic and Molecular Bases of Inherited Disease New York, McGraw-Hill; 2001. Vol III, pp4933–4956. [Google Scholar]
- Mykkanen J, Torrents D, Pineda M, et al. Functional analysis of novel mutations in y(+)LAT-1 amino acid transporter gene causing lysinuric protein intolerance (LPI) Hum Mol Genet. 2000;9:431–438. doi: 10.1093/hmg/9.3.431. [DOI] [PubMed] [Google Scholar]
- Sperandeo MP, Bassi MT, Riboni M, et al. Structure of the SLC7A7 gene and mutational analysis of patients affected by lysinuric protein intolerance. Am J Hum Genet. 2000;66:92–99. doi: 10.1086/302700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A, Shoji Y, Koizumi A, et al. SLC7A7 genomic structure and novel variants in three Japanese lysinuric protein intolerance families. Hum Mutat. 2000;15:367–372. doi: 10.1002/(SICI)1098-1004(200004)15:4<367::AID-HUMU9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Cimbalistiene L, Lehnert W, Huoponen K, Kucinskas V. First reported case of lysinuric protein intolerance (LPI) in Lithuania, confirmed biochemically and by DNA analysis. J Appl Genet. 2007;48:277–280. doi: 10.1007/BF03195224. [DOI] [PubMed] [Google Scholar]
- Sperandeo MP, Andria G, Sebastio G. Lysinuric protein intolerance: update and extended mutation analysis of the SLC7A7 gene. Hum Mutat. 2008;29:14–21. doi: 10.1002/humu.20589. [DOI] [PubMed] [Google Scholar]
- Palacín M, Nunes V, Font-Llitjós M, et al. The genetics of heteromeric amino acid transporters. Physiology (Bethesda) 2005;20:112–124. doi: 10.1152/physiol.00051.2004. [DOI] [PubMed] [Google Scholar]
- Tanner LM, Nanto-Salonen K, Niinikoski H, et al. Nephropathy advancing to end-stage renal disease: a novel complication of lysinuric protein intolerance J Pediatr 2007150631–634.634.e1. [DOI] [PubMed] [Google Scholar]
- Palacin M, Bertran J, Chillaron J, Estevez R, Zorzano A. Lysinuric protein intolerance: mechanisms of pathophysiology. Mol Genet Metab. 2004;81 Suppl 1:S27–S37. doi: 10.1016/j.ymgme.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Broer S. Lysinuric protein intolerance: one gene – many problems. Am J Physiol Cell Physiol. 2007;293:C540–C541. doi: 10.1152/ajpcell.00166.2007. [DOI] [PubMed] [Google Scholar]
- Tanner LM, Nanto-Salonen K, Niinikoski H, Huoponen K, Simell O. Long-term oral lysine supplementation in lysinuric protein intolerance. Metabolism. 2007;56:185–189. doi: 10.1016/j.metabol.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lauteala T, Mykkanen J, Sperandeo MP, et al. Genetic homogeneity of lysinuric protein intolerance. Eur J Hum Genet. 1998;6:612–615. doi: 10.1038/sj.ejhg.5200236. [DOI] [PubMed] [Google Scholar]
- Lauteala T, Sistonen P, Savontaus ML, et al. Lysinuric protein intolerance (LPI) gene maps to the long arm of chromosome 14. Am J Hum Genet. 1997;60:1479–1486. doi: 10.1086/515457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents D, Mykkanen J, Pineda M, et al. Identification of SLC7A7, encoding y+LAT-1, as the lysinuric protein intolerance gene. Nat Genet. 1999;21:293–296. doi: 10.1038/6809. [DOI] [PubMed] [Google Scholar]
- Borsani G, Bassi MT, Sperandeo MP, et al. SLC7A7, encoding a putative permease-related protein, is mutated in patients with lysinuric protein intolerance. Nat Genet. 1999;21:297–301. doi: 10.1038/6815. [DOI] [PubMed] [Google Scholar]
- Puomila K, Simell O, Huoponen K, Mykkanen J. Two alternative promoters regulate the expression of lysinuric protein intolerance gene SLC7A7. Mol Genet Metab. 2007;90:298–306. doi: 10.1016/j.ymgme.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Torrents D, Estévez R, Pineda M, et al. Identification and characterization of a membrane protein (y+L amino acid transporter-1) that associates with 4F2hc to encode the amino acid transport activity y+L. A candidate gene for lysinuric protein intolerance. J Biol Chem. 1998;273:32437–32445. doi: 10.1074/jbc.273.49.32437. [DOI] [PubMed] [Google Scholar]
- Palacín M, Borsani G, Sebastio G. The molecular bases of cystinuria and lysinuric protein intolerance. Curr Opin Genet Dev. 2001;11:328–335. doi: 10.1016/s0959-437x(00)00198-2. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, Choo KH. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA) J Med Genet. 2003;40:907–912. doi: 10.1136/jmg.40.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez L, Garcia-Cazorla A, Gutierrez A, et al. Treatment of severe osteoporosis with alendronate in a patient with lysinuric protein intolerance. J Inherit Metab Dis. 2006;29:687. doi: 10.1007/s10545-006-0236-9. [DOI] [PubMed] [Google Scholar]
- Tsumura H, Suzuki N, Saito H, et al. The targeted disruption of the CD98 gene results in embryonic lethality. Biochem Biophys Res Commun. 2003;308:847–851. doi: 10.1016/s0006-291x(03)01473-6. [DOI] [PubMed] [Google Scholar]
- Fort J, de la Ballina LR, Burghardt HE, et al. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J Biol Chem. 2007;282:31444–31452. doi: 10.1074/jbc.M704524200. [DOI] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- Reig N, del Rio C, Casagrande F, et al. Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: a model for APC transporters. J Biol Chem. 2007;282:13270–13281. doi: 10.1074/jbc.M610695200. [DOI] [PubMed] [Google Scholar]
- Stiburek L, Fornuskova D, Wenchich L, Pejznochova M, Hansikova H, Zeman J. Knockdown of human Oxa1l Impairs the biogenesis of F(1)F(o)-ATP synthase and NADH:Ubiquinone oxidoreductase. J Mol Biol. 2007;374:506–516. doi: 10.1016/j.jmb.2007.09.044. [DOI] [PubMed] [Google Scholar]
- Purandare SM, Patel PI. Recombination hot spots and human disease. Genome Res. 1997;7:773–786. doi: 10.1101/gr.7.8.773. [DOI] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- Kanno J, Hutchin T, Kamada F, et al. Genomic deletion within GLDC is a major cause of non-ketotic hyperglycinaemia. J Med Genet. 2007;44:e69. doi: 10.1136/jmg.2006.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Willett-Brozick JE, Filho PA, Lawrence EC, Myers EN, Ferrell RE. An Alu-mediated partial SDHC deletion causes familial and sporadic paraganglioma. J Med Genet. 2004;41:703–709. doi: 10.1136/jmg.2004.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Jaworski A, Larson JE, et al. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.