Abstract
The introduction of prenatal screening requires rapid high-throughput diagnosis of common aneuploidies. Multiplex ligation-dependent probe amplification (MLPA) allows for quick, easily automated multiplex testing of these aneuploidies in one polymerase chain reaction. We performed a large prospective study using MLPA on 4000 amniotic fluid (AF) samples including all indications and compared its value to karyotyping and fluorescence in situ hybridization (FISH). MLPA can reliably determine common aneuploidies with 100% sensitivity and 100% specificity. Moreover, some mosaic cases and structural chromosome aberrations were detected as well. In cases of a male fetus, triploidies can be detected by an aberrant pattern of probe signals, which mimics maternal cell contamination (MCC). Macroscopic blood contamination was encountered in 3.2% of the AF samples. In 20% of these samples, an MLPA pattern was found consistent with MCC, although there were no false negatives of the most common aneuploidies. As the vast majority of inconclusive results (1.7%) is due to potential MCC, we designed a protocol in which we determine whether MLPA can be performed on blood-contaminated AF samples by testing if blood is of fetal origin. Then, the number of inconclusive results could be theoretically reduced to 0.05%. We propose an alternative interpretation of relative probe signals for rapid aneuploidy diagnosis (RAD). We discuss the value of MLPA for the detection of (submicroscopic) structural chromosome anomalies. MLPA is a reliable method that can replace FISH and could be used as a stand-alone test for RAD instead of karyotyping.
Keywords: MLPA, rapid aneuploidy diagnosis, chromosomal mosaicism, maternal cell contamination, copy number variations
Introduction
In addition to karyotyping, the possibility for quick, targeted diagnosis of a selection of chromosome anomalies is expanding. Fluorescence in situ hybridization (FISH) on uncultured amniotic fluid (AF) cells has been proved to be a reliable technique for the rapid detection of the most common aneuploidies rapid aneuploidy diagnosis (RAD). Moreover, it has a major additional diagnostic value in the further characterization of chromosome anomalies found by karyotyping.1 Furthermore, in the case of a male fetus, maternal cell contamination (MCC) in uncultured AF samples can be demonstrated.2, 3, 4 However, the technique is labor-intensive and not easily applicable to automatic handling of a large number of samples.
A valuable alternative is quantitative fluorescent-polymerase chain reaction (QF-PCR), which is less expensive and more suitable for automation. It has been diagnostically validated for the use of simultaneous testing of up to 12 highly polymorphic short tandem repeat markers on loci pre-selected on certain chromosomes of choice.5, 6, 7 It can reliably detect MCC in AF of both male and female fetuses. A small percentage (0.08%) of non-informative results, depending on the number of markers per chromosome, have been reported.6 When handling over 1100 samples a year, it is a cost-effective alternative to karyotyping for rapid prenatal diagnosis of common aneuploidies.8
A comparable but more recently developed PCR-based technique named multiplex ligation-dependent probe amplification (MLPA) allows for relative quantification of up to 45 DNA target sequences in one PCR.9 It does not require living cells or cell culture, but the input of 20 ng or more DNA. It is less labor-intensive, can be better automated and is less expensive as compared with karyotyping and FISH. Its turn around time can be as quick as 30 h. MLPA has been used prospectively for prenatal diagnosis on 784 CVS and 809 AF samples.10 To be truly validated as a diagnostic test, there is a call for larger studies.11, 12 To assess whether MLPA is a reliable tool for the detection of the most common aneuploidies, we performed MLPA on 4000 AF samples in addition to karyotyping.
Here, we report on MLPA test results on 4000 AF samples taken for all indications, discuss the influence of MCC and propose a protocol for handling blood-contaminated AF samples. We present a standard procedure for the detection of common aneuploidies and chromosomal mosaicism. We also discuss the value of MLPA compared with FISH in RAD, its potential to detect (submicroscopic) structural chromosome anomalies and its value as a stand-alone test in prenatal diagnosis.
Materials and methods
MLPA reaction
The principle of MLPA is extensively described.9 Briefly, a set of two probes is designed to hybridize adjacent to each other on one DNA target sequence. For the SALSA MLPA P095 aneuploidy kit, eight target sequences are chosen for each of the chromosomes 13, 18, 21 and X. The Y chromosome has four target sequences. The probes consist of a target sequence and a universal forward or reverse PCR primer-binding sites. In between these, one of the probes contains the so-called stuffer sequence of a specific length, varying from 130 to 490 bp (base pairs). This length is specific for a target sequence. After hybridization, the probes are ligated and PCR is performed with a universal fluorescent-labeled primer pair. The relative amount of PCR product is proportional to the amount of target sequence. The amplification products of different lengths are separated by sequence-type electrophoresis.
Samples
From May 2004 until August 2007, we received 4911 AF samples in our laboratory for cytogenetic diagnosis. The referral reasons covered the whole range of prenatal indications for invasive testing. We prospectively investigated 4000 AF samples with the SALSA MLPA P095 aneuploidy kit from MRC-Holland BV (Amsterdam, The Netherlands), and the outcomes were compared with the karyotyping results of the cell cultures. In the remaining 911 cases, MLPA was not performed because of an insufficient amount of available AF sample for MLPA testing in most cases. In 505 out of 4000 cases, MLPA outcomes were also compared with FISH results for RAD.
Once a week MLPA was performed routinely on nearly all samples that were received during the week before. In general, tests were processed in batches of about 30 samples including two normal male and one normal female controls and a blank. Data processing of the MLPA tests was performed without the knowledge of karyotype or FISH results. At the end of the study period, DNA isolation from both AF tubes of each patient and the MLPA reaction and analysis were performed in duplicate by two technicians. Results were reported only if the conclusions of both tubes match.
Sample preparation
DNA was isolated from 1 to 5 ml AF, with an average of 2 ml, depending on the total amount of AF received. In the beginning of this study, DNA was isolated using the QIAamp DNA Mini Kit from Qiagen (Hilden, Germany) according to the manufacturer's instructions (n∼1000). Later the ABI Prism 6100 Nucleic Acid PrepStation (Applied Biosystems, Foster City, CA, USA) was used (n∼1800), and since August 2006, DNA is isolated with the Chemagic Magnetic Separation Module I (Chemagen, Baesweiler, Germany) (n∼2200) with 96 needles allowing the isolation of 96 samples at the same time which makes it less labor- and time-intensive.
DNA yield is in the range of 2–10 ng/μl with a total volume of 50 μl. Routinely we use 7 μl (namely 14–70 ng) of the DNA solution per MLPA reaction. MLPA reactions are performed on a PCR thermocycler with heated lid (Biometra Thermal Cycler, Westburg, The Netherlands). We have adapted the protocol of MRC-Holland, in that we used all of the ligation mixture in the PCR reaction. 2μl of the PCR product was analyzed by capillary electrophoresis on a ABI 3100 GeneScan in the beginning of the study (until august 2006) (n=2752) and later on a ABI 3730 GeneScan (n=1248) (both from Applied Biosystems).
Analysis
Data analysis is performed using GeneMarker 1.51 software (SoftGenetics, LLC, State College, PA, USA). The size and peak area are analyzed for each MLPA probe. The calculation of relative probe signals is performed on samples processed within an assay run. In chromosomally normal samples, the relative probe signal is expected to be 1 for all probes on the autosomes and the sex chromosomes if, for the latter, samples of the same sex are set as controls. Routinely we set normal male samples as controls (Figure 1). A chromosome aberration is better visible if normal samples of the same sex as that from the fetus are set as controls (Figure 2A and B).
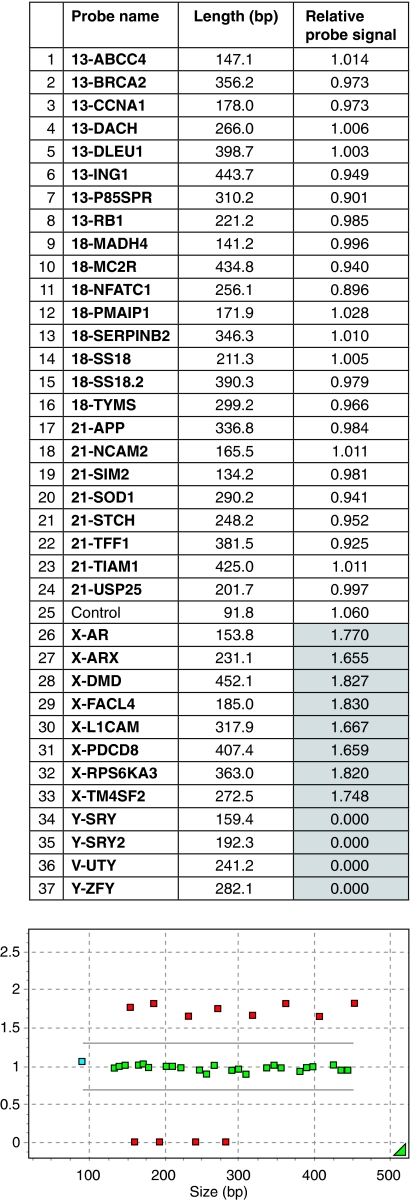
Figure 1.
This is a part of the MLPA analysis report (Genemarker v1.51) of a normal female. In the upper part, all 37 probes in the SALSA MLPA P095 aneuploidy kit are shown in the second column with their length in base pairs in the third column. The last column represents the relative probe signal of each probe. Normal values are defined between 0.7 and 1.3 and are shown in a white background. If the relative probe signal is aberrant, it has a gray background. In this case, a normal male is set as a control, hence the X and Y probes are aberrant; there are no relative probe signals for Y and the relative probe signals for X are nearly 2. The lower part of the figure shows the length in base pairs on the X axes and the relative probe signal on the Y axes. Normal relative probe signals are between the grey lines (0.7 and 1.3) and are depicted in green. Aberrant relative probe signals are depicted in red.
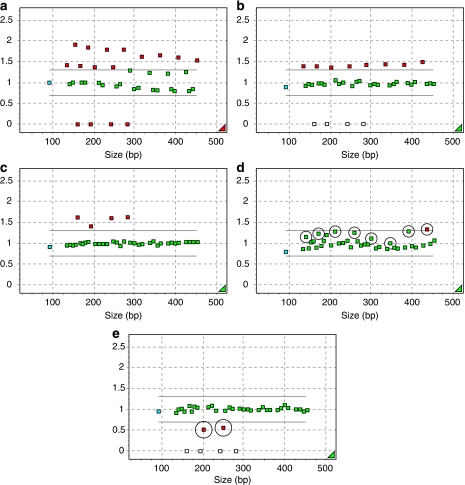
Figure 2.
(a) Part of an MLPA analysis report (Genemarker v1.51) of a trisomy 21 case: when normal samples of the opposite sex as that of the fetus are used as controls only 4 of 8 probes are >1.3. (b) Part of an MLPA analysis report (Genemarker v1.51) of the same trisomy 21 case as in (a) when normal samples of the same sex as that of the fetus are used as controls 8 of 8 probes are >1.3. (c) Part of an MLPA analysis report (Genemarker v1.51) of the XYY/XY mosaic: the relative probe signals of all Y probes are > 1.3. (d) Part of an MLPA analysis report (Genemarker v1.51) of the mosaic trisomy 18: the relative probe signals of 6 of 8 chromosome 18 probes are >cutoff values (see Table 3). (e) Part of an MLPA analysis report (Genemarker v1.51) of a partial deletion of chromosome 21, which showed to be an unbalanced complex translocation of an inverted chromosome 4 with chromosome 21. Normal females were set as controls.
To enable the detection of chromosomal mosaicism, we calculated cutoff values for different probes on different chromosomes for males and females. These were calculated by considering the median probe signal±2 × SD (standard deviation) using 667 normal male samples and 694 normal female samples.
To reduce the number of inconclusive results, we have designed a protocol in which we selectively handle and issue an MLPA result of macroscopic blood-contaminated AF samples. First, we determined the fetal hemoglobin (HbF) level in AF. Briefly, the percentage of HbF of total Hb, including adult Hb, is determined by the alkali denaturation method.13 After adding NaOH, there is a difference in the decrease of absorbance of Hb as measured by spectophotometry between HbF and adult Hb. If the HbF level is ≥85% of the total Hb level, we consider the blood of fetal origin (WJ Kleijer, unpublished results) and perform MLPA. If the fetal Hb level is lower than 85%, we consider the sample to be contaminated with maternal blood and perform FISH.
For cytogenetic analysis, AF cells were cultured according to the standard techniques on culture slides (BD Falcon, Bedford, MA, USA) and cultures were harvested using the in situ method. GTG banding was used for chromosome analysis and metaphases of 10 cell colonies were routinely investigated. FISH on uncultured AF cells for RAD was performed as described earlier.14, 15
Results
MLPA compared with karyotyping
Four thousand AF samples were prospectively investigated with MLPA with the P095 aneuploidy kit and compared with karyotyping. In 230 of 4000 samples (5.8%), chromosome aberrations were found, from which 140 (61%) samples represent the most common aneuploidies.
There were 3932 conclusive (98.3%) and 68 (1.7%) inconclusive results (Table 1).
Table 1. Summary of the results of the prospective study on 4000 AF samples.
| MLPA result | n | Chromosome result | n |
|---|---|---|---|
| Conclusive | (3932) | ||
| Normal male/female | 3783 | Normal 46,XY or 46,XX | 3707 |
| Abnormal | 76 | ||
| 69,XXX | 4 | ||
| mos 47,XXY[5]/(47,XXY/46,XY)[1]/46,XY[45] | 1 | ||
| mos 45,X[5]/(45,X/46,XY)[1]/46,XY[20] | 1 | ||
| (mos) 47,+ mar | 8 | ||
| (mos) aneuploidy (no 13, 18, 21, X, Y) | 6 | ||
| Structural chromosome aberration | 56 | ||
| Abnormal | |||
| 1. Common aneuploidies | |||
| Trisomy 21 | 75 | 47,XX or XY,+21 | 75 |
| Trisomy 18 | 24 | 47,XX or XY,+18 | 24 |
| Trisomy 13 | 17 | 47,XX or XY,+13 | 16 |
| 46,XX,+13,der(13;14)(q10;q10)pat | 1 | ||
| Monosomy X | 8 | 45,X | 8 |
| Triple X | 3 | 47,XXX | 3 |
| XXY | 8 | 47,XXY | 8 |
| XYY | 1 | 47,XYY | 1 |
| Triploidy XXY | 3 | 69,XXY | 3 |
| 2. Other aberrations | |||
| 2a. Mosaics | |||
| mos XXY/XY | 1 | mos 47,XXY[6]/46,XY[21] (22%) | 1 |
| mos XYY/XY | 1 | mos 45,X[2]/46,XY[12] (14%)a | 1 |
| mos X/XY | 3 | mos 45,X[63]/46,XY[2] (97%) | 1 |
| mos 45,X[12]/46,XY[8] (60%) | 1 | ||
| mos 45,X[15]/(45,X/46,XY)[3]/46,XY[25] (39%) | 1 | ||
| mos XY,+21 | 1 | mos 47,XY,+21[8]/46,XY[25] (24%) | 1 |
| mos XY,+18 | 1 | 47,XY,+18b | 1 |
| 2b. Structural chromosome aberrations | |||
| amp(13) (CCNA) | 1 | 46,XX,der(17)ins(17;13)(q11.2;q12.3q14.1)mat | 1 |
| del(21) (USP25, STCH) | 1 | 45,XX,der(4)inv(4)(p16q35)t(4;21)(q35;q11),-21 | 1 |
| del(18) (TYMS) | 1 | 46,XX.ish del(18)(p11.32p11.32)(RP11-145B19-)c | 1 |
| Inconclusive | (68) | ||
| Potential MCC | 58 | 46,XX | 51 |
| 46,XY | 3 | ||
| mos 47,XX,+2[11]/46,XX[14] (44%) | 1 | ||
| 69,XXX | 1 | ||
| 46,XX,inv(13)(q14.2q21.1)pat | 1 | ||
| 46,XX,t(9;20)(q32;p13)pat | 1 | ||
| Insufficient amount of DNA | 8 | 46,XX | 3 |
| 46,XY | 4 | ||
| 47,XXY | 1 | ||
| Unknown reasons | 2 | 46,XY | 2 |
| Total | 4000 | 4000 |
The mosaic XYY/XY with MLPA and QF-PCR showed to be a mosaic 45,X/46,XY in the cell cultures. An explanation might be that both abnormal cell lines are the products of non-disjunction of an XY cell during early embryonic development with selection against the XYY cell line in AF cell cultures.
With FISH on uncultured AF cells, a mosaic trisomy 18 was found as well, with 26% of the cells showing three signals. The karyotype was 47,XY,+18 in 61 cell clones.
The deletion was confirmed with FISH with an overlapping BAC clone RP11-145B19.
Conclusive results
Of the 3783 samples with a normal MLPA result, karyotyping of the cell cultures revealed a normal karyotype in 3707 (98.1%) cases and a chromosome aberration other than the most common aneuploidies in 76 (1.9%) cases; the MLPA kit was not expected to detect these cases. This includes 56 cases with a structural chromosome aberration, four cases of 69,XXX, two sex-chromosomal mosaic cases, six samples with (mosaic) aneuploidies other than the most common ones and eight samples with mosaicism of an extra marker chromosome (manuscript in preparation).
Of the 149 cases with an abnormal MLPA result, 116 cases with a trisomy of either chromosome 13, 18 or 21 were found. In 113 out of 116 samples, the relative probe signals of at least five probes were ≥1.3. In the remaining three cases, only one or two relative probe signals were ≥1.3, but these AF samples were blood contaminated, and therefore potentially contaminated with maternal cells (see Table 2). Among 129 blood-contaminated samples, 7 trisomies were present, which were all detected with MLPA (Table 2). In one case, trisomy 13 showed to be the result of an unbalanced familial Robertsonian translocation der(13;14)(q10;q10). In 20 cases, MLPA results were indicative for monosomy X (n=8), triple X (n=3), XXY (n=8) or XYY (n=1). All were confirmed with classical chromosome analysis.
Table 2. MLPA results of 129 blood-contaminated AF samples.
| MLPA result | n | n probes >1.3 | n probes >cutoff value | Interpretation | Karyotype (n cases) |
|---|---|---|---|---|---|
| Conclusive | (71 cases) | ||||
| Normal XY | 44 | Normal XY | 46,XY (n=43) | ||
| 46,XY,der(4),t(5;9)(p13;p22) (n=1) | |||||
| XY/XX | 20 | Normal XY with MCC | 46,XY (n=19) | ||
| 46,XY,del(13)(q22;q31) (n=1) | |||||
| XY,+21 | 1 | 8 | 8 | XY,+21 | 47,XY,+21 |
| XY,+21 | 1 | 6 | 8 | XY,+21 | 47,XY,+21 |
| XY/XX,+21 | 1 | 2 | 8 | XY,+21 with MCC | 47,XY,+21 |
| XY/XX,+18 | 1 | 1 | 6 | XY,+18 with MCC | 47,XY,+18 |
| XX,+18 | 1 | 8 | 8 | XX,+18 | 47,XX,+18 |
| XX,+18 | 1 | 7 | 8 | XX,+18 | 47,XX,+18 |
| XX,+18 | 1 | 2 | 8 | XX,+18 with MCC | 47,XX,+18 |
| Inconclusive | (58 cases) | ||||
| Normal XX | 58 | Inconclusive | See Table 1 | ||
| Total | 129 |
In three male cases, MLPA results were abnormal, suggestive of MCC or triploidy. As all three AF samples were without blood contamination and fetal ultrasound (US) abnormalities were suggestive of triploidy, MLPA results could be interpreted as such, however, with caution because MCC of a normal male sample was not really excluded.
Although the MLPA P095 kit is designed for the detection of the common aneuploidies, some other chromosome aberrations, like chromosomal mosaicism and structural chromosome aberrations, were detected as well (see Table 1). To be able to detect mosaicism, we calculated cutoff values for all the probes in the P095 MLPA kit (shown in Table 3). We found five sex-chromosomal and two autosomal mosaicisms. In the first case mentioned in Table 1, the relative probe signals of seven X-probes were elevated as compared with the normal cutoff values. This showed to be a mosaic 47,XXY[6]/46,XY[21] (mosaicism of 22%). In the second case, a mosaic XYY/XY was found with the relative probe signals of all Y-probes >1.3, but <2 as expected in a full-blown XYY (Figure 2c). Surprisingly, a mosaic 45,X[2]/46,XY[12] was found in the cell cultures. The MLPA result was corroborated by QF-PCR of the SRY gene and by PCR of the Amelogenin Y gene (data not shown).
Table 3. Cutoff values for the different probes on the different chromosomes: median probe signal±2 × SD (normal males were set as controls).
| Cutoff value female (N=694) | Cutoff value male (N=667) | |||
|---|---|---|---|---|
| Probe | Minimum | Maximum | Minimum | Maximum |
| 21-SIM2 | 0.837 | 1.105 | 0.863 | 1.125 |
| 21-NCAM2 | 0.828 | 1.102 | 0.841 | 1.135 |
| 21-USP25 | 0.881 | 1.079 | 0.884 | 1.100 |
| 21-STCH | 0.820 | 1.150 | 0.841 | 1.169 |
| 21-SOD1 | 0.805 | 1.095 | 0.848 | 1.142 |
| 21-APP | 0.812 | 1.108 | 0.871 | 1.145 |
| 21-TFF1 | 0.830 | 1.096 | 0.873 | 1.150 |
| 21-TIAM1 | 0.775 | 1.109 | 0.841 | 1.147 |
| 13-ABCC4 | 0.877 | 1.094 | 0.878 | 1.110 |
| 13-CCNA1 | 0.838 | 1.178 | 0.843 | 1.199 |
| 13-RB1 | 0.888 | 1.096 | 0.904 | 1.122 |
| 13-DACH | 0.813 | 1.121 | 0.851 | 1.149 |
| 13-P85SPR | 0.839 | 1.121 | 0.885 | 1.147 |
| 13-BRCA2 | 0.837 | 1.123 | 0.869 | 1.151 |
| 13-DLEU1 | 0.757 | 1.104 | 0.836 | 1.151 |
| 13-ING1 | 0.803 | 1.131 | 0.859 | 1.185 |
| 18-MADH4 | 0.866 | 1.072 | 0.893 | 1.093 |
| 18-PMAIP1 | 0.856 | 1.120 | 0.894 | 1.124 |
| 18-SS18 | 0.848 | 1.100 | 0.871 | 1.131 |
| 18-NFATC1 | 0.829 | 1.115 | 0.857 | 1.145 |
| 18-TYMS | 0.836 | 1.082 | 0.875 | 1.113 |
| 18-SERPINB2 | 0.766 | 1.137 | 0.811 | 1.184 |
| 18-SS18,2 | 0.825 | 1.107 | 0.847 | 1.153 |
| 18-MC2R | 0.782 | 1.130 | 0.832 | 1.163 |
| X-AR | 1.589 | 2.138 | 0.807 | 1.193 |
| X-ARX | 1.543 | 2.094 | 0.825 | 1.201 |
| X-DMD | 1.420 | 2.107 | 0.755 | 1.165 |
| X-FACL4 | 1.468 | 2.126 | 0.775 | 1.213 |
| X-L1CAM | 1.456 | 2.076 | 0.781 | 1.219 |
| X-PDCD8 | 1.438 | 2.088 | 0.807 | 1.193 |
| X-RPS6KA3 | 1.485 | 1.995 | 0.819 | 1.179 |
| X-TM4SF2 | 1.482 | 2.025 | 0.782 | 1.214 |
| Y-SRY | 0 | 0 | 0.769 | 1.255 |
| Y-SRY2 | 0 | 0 | 0.631 | 1.257 |
| Y-UTY | 0 | 0 | 0.747 | 1.253 |
| Y-ZFY | 0 | 0 | 0.825 | 1.225 |
Three cases of 45,X/46,XY mosaicism (39, 60 and 97%) were correctly identified, as in the first two cases the relative probe signals of all Y-probes were decreased as compared with the normal cutoff values. In the third case, Y-signals were seen for two of the four Y-probes. Besides these sex chromosomal mosaics, there were two autosomal mosaics. A mosaic trisomy 21 was identified, as the relative probe signals of 4 of 8 probes were above the normal cutoff values, matching a 24% mosaicism in cell culture. A case of trisomy 18 mosaicism (relative probe signals of 6 of 8 probes were above the normal cutoff values) confirmed by FISH analysis on uncultured AF cells in 26% of the interphase nuclei, surprisingly, showed to be a full-blown trisomy 18 in the cell cultures (61 cell clones) (Figure 2d).
On the basis of the MLPA results, a structural autosomal aberration was suspected in three samples because one or two probes on a specific chromosome showed (an) abnormal relative probe signal(s):
Case 1: An amplification of the CCNA probe on chromosome 13q12.3 showing to be an unbalanced maternal insertion of 13q12.3–q14.1 into 17q11.2.
Case 2: A deletion of the 21q11 probes (USP25 on 21q11.2 and STCH on 21q11) due to a complex unbalanced translocation t(4;21) (Figure 2e).
Case 3: A submicroscopic deletion of TYMS on chromosome 18p11.3, which was confirmed with FISH with an overlapping BAC clone.
Inconclusive results
In 68 cases (1.7%), results were inconclusive (see Table 1) due to:
Blood contamination of the AF itself or the cell pellet after centrifugation and therefore potential MCC (n=58). In these cases, a normal female MLPA profile was found. Three of these cases showed a normal male karyotype, interpreted as MCC, and four cases had a female karyotype with a chromosome aberration undetectable by MLPA (Table 1).
An insufficient amount of DNA as indicated by the DNA quantity control fragments in the kit (for details see the MRC-Holland website) (n=8). In all but one case of Klinefelter's syndrome, a normal karyotype was found.
Unknown reasons (n=2). Both showed a normal male karyotype.
MLPA compared with FISH
During the study period, FISH analysis was the golden standard for RAD of the most common aneuploidies. To determine whether MLPA can replace FISH for RAD, we compared FISH and MLPA results in 505 out of 4000 samples (Table 4). In all these cases, there was an indication for RAD, for instance in the case of certain fetal US abnormalities approaching 24 weeks of gestational age. In The Netherlands, termination of pregnancy is generally allowed up to 24 weeks of gestation.
Table 4. MLPA and FISH results in 505 AF samples.
| MLPA/FISH results | N |
|---|---|
| Concordant results | 495 |
| Normal XX | 197 |
| Normal XY | 241 |
| Trisomy 13 | 11 |
| Trisomy 18 | 8 |
| Trisomy 21 | 28 |
| Triploidy XXY | 2 |
| XXX | 2 |
| XXY | 2 |
| Monosomy X | 2 |
| Mos X/XY (97% with FISH) | 1 (45,X[63]/46,XY[2]) |
| Mos trisomy 18 (26% with FISH) | 1a |
| Discordant results | 10 (see Table 5) |
| Total | 505 |
A 100% trisomy 18 (61 cell clones) was found in the AF cell cultures.
In 495 (98%) out of 505 samples, concordant FISH and MLPA results were achieved, which were all confirmed by karyotyping of the cell cultures, although in one case a mosaic trisomy 18 was suspected on the basis of FISH and MLPA results, whereas the cell cultures revealed a 100% trisomy 18 (Figure 2d). Ten cases (2%) revealed discordant results (Table 5). In two cases a triploidy (69,XXX) was involved, which, because of the nature of the technique, is not detectable with MLPA. FISH was able to detect low-level mosaicism such as in case 5, whereas FISH was also responsible for some potential false-positive mosaic cases (cases 6, 9 and 10). However, on the basis of the investigated number of cell clones, low-level mosaicism in the cell cultures cannot be excluded (see comments in Table 5). Both cases of XY contamination of an XX sample (cases 7 and 8) as revealed by FISH were unexplained contaminations. The triple X cell line seen with FISH in case 3, although not seen in the AF cell cultures, was confirmed in fetal skin. The normal cell line as detected with FISH in case 4 was neither confirmed in AF cell cultures nor in the skin, amnion and placenta.
Table 5. Discordant MLPA and FISH results and corresponding karyotype.
| Case | MLPA result | FISH result (number of nuclei) | Karyotype (number of cell clones) | Comments |
|---|---|---|---|---|
| 1 | Normal XX | Triploidy XXX [50] | 69,XXX [5] | 69,XXX not detectable with MLPA |
| 2 | Normal XX | Triploidy XXX [50] | 69,XXX [5] | 69,XXX not detectable with MLPA |
| 3 | Monosomy X | Mos X(90%)/XXX(10%) [100] | 45,X [41] | 41 cell clones exclude mosaicism of ≥8%a |
| Mosaicism confirmed in skin fibroblasts X(80%)/XXX (17%)/XX (3%) | ||||
| 4 | Monosomy X | Mos X(85%)/46,XY(15%) [40] | 45,X [37] | 37 cell clones exclude mosaicism of ≥8%a |
| 100% 45,X confirmed in the skin, amnion and placenta | ||||
| 5 | Normal XY | Mos X(7%)/XY(93%) [100] | 45,X[5]/(45,X/46,XY) [1]/46,XY [20] | |
| 6 | Normal XX | Mos XXX(12%)/XX(88%) [260] | 46,XX [26] | 26 cell clones exclude mos of ≥11%a |
| 7 | Normal XX | Mos XX(90%)/XY(10%) [50] | 46,XX [31] | 31 cell clones exclude mos of ≥10%a |
| 8 | Normal XX | Mos XX(93%)/46,XY(7%) [100] | 46,XX [16] | 16 cell clones exclude mos of ≥18%a |
| 9 | Normal XY | Mos XXY(5%)/XY(95%) [300] | 46,XY [16] | 16 cell clones exclude mos of ≥18%a |
| 10 | Normal XY | Mos XYY(6%)/XY(94%) [200] | 46,XY [22] | 22 cell clones exclude mos of ≥13%a |
See Hook (1977).23
Discussion
According to the manufacturer's instructions, a trisomy is indicated by a relative probe signal ≥1. 3 and a monosomy by a relative probe signal of ≤0.7. On the basis of our results, we consider the presence of a trisomy if at least 4 of 8 probes are ≥1.3, and the relative probe signals of the remaining four probes are above the cutoff values (see Table 3). If normal males are set as controls, we consider the presence of:
A monosomy X if the relative probe signals of probes on the X-chromosome are within the normal ranges of those for normal males (see Table 3) and Y-signals are absent.
An XXY if the relative probe signals of the Y-probes are within the normal ranges of those for normal males and those of the X-probes are within the normal ranges of those from normal females (see Table 3).
An XYY if the relative probe signals of the Y-probes are ∼1.8 and those of the X-probes are within the normal ranges of those from normal males (see Table 3).
An XXX if the relative probe signals of the X-probes are ∼2.5 and those of the Y-probes are 0.
We have shown that MLPA can indeed reliably determine the most common aneuploidies with 100% sensitivity and 100% specificity. It has been reported that triploidies11, 16 or 69,XXX only10, 12, 17 cannot be diagnosed by MLPA. However, if an AF sample is not blood contaminated, we show that in cases of a male fetus, triploidies can be detected by an aberrant pattern of MLPA probe signals, which mimics MCC. A female fetus with triploidy can indeed not be diagnosed by MLPA. On fetal US examination, only about 80% of the cases can be detected;18 hence, theoretically up to about 10% of female triploidy cases will potentially be missed using combined MLPA and fetal US examination. Although most of the cases of triploidies will end in intrauterine fetal death, the obstetric management can be hampered by lack of information on the fetal karyotype. In our series, we found 8 of 4000 (0.2%) cases of triploidy for all indications. Therefore, we recommend using FISH or QF-PCR for genotyping when there is a suspicion of triploidy after fetal US examination, especially if the sex of the fetus is female or unknown.
In prenatal genotyping, potential misdiagnosis of aneuploidies due to MCC is a major concern. Visual inspection of the AF samples or the cell pellet after centrifugation does not exclude MCC.2, 4 It has been reported that blood-contaminated samples cannot be processed by MLPA, or that MCC cannot be diagnosed by MLPA.11, 12, 20 Before determining the effects of MCC on MLPA results, we established the relative occurrence of MCC in our AF samples. We did so because this has been reported to differ between laboratories, due to the invasive procedure itself, which is a significant contributor to MCC.19, 20 It has been shown that MCC can be diagnosed in uncultured AF samples with FISH by determining the number of AF cells with two chromosome X signals in a male fetus.2 We have used this FISH procedure on 447 AF samples in a high-risk population and found that in 2.2% of the samples MCC of 10% or more is present. These samples all were macroscopically blood contaminated. Therefore, MCC of more than 10% can be pre-selected by the visual inspection of AF sample. However, this cannot always be performed for MCC lower than 10%. In addition, MCC tests, using selective testing of highly polymorphic markers by QF-PCR, hardly diagnose this low-grade MCC.21 We decided to do the MLPA test on all AF samples including those with macroscopic blood contamination to be able to determine whether MCC does affect the results of MLPA RAD testing.
In our series of 4000 MLPA cases, we had 129 cases of macroscopic blood-contaminated AF samples (3.2%). In seven cases, a trisomy 18 (n=4) or trisomy 21 (n=3) was present, which were all detected with MLPA, although MCC was seen in three of these cases. In 20 out of 64 normal male cases, we found an MLPA profile consistent with a majority of XY cells and a minority of XX cells. The XX contamination was interpreted as MCC, as true fetal chimerism XX/XY is very rare indeed.22 In 58 cases, we found a normal MLPA profile consistent with a female fetus. These results were interpreted as inconclusive due to potential MCC. In three of these cases, a normal male karyotype was found, which is most likely explained by substantial MCC. Overall, an MLPA profile consistent with MCC was found in 26 of 129 (20%) of the blood-contaminated samples. In 55 of 129 (43%) samples, potential MCC was present. To reduce the number of inconclusive results, we have designed a protocol in which we selectively determine whether MLPA can be performed by testing if the blood is of fetal origin. If so, we perform MLPA. If not, we perform FISH because in our laboratory a detection rate of about 5% of cells with a numerical chromosome anomaly can be achieved with FISH by increasing the number of analyzed cells (normally 50 cells per probe). In case of normal female signal distributions, karyotyping is needed for a reliable diagnosis. Overall, we did not encounter false negatives using MLPA of the most common aneuploidies, which could be attributed to MCC. Theoretically, we can reduce the number of inconclusive MLPA results to 0.05% by using the measurement of HbF in blood-contaminated samples and by excluding samples with insufficient amount of DNA.
Chromosomal mosaicism is generally excluded up to 26% by analyzing 10 cell clones by karyotyping.23 Interphase FISH, QF-PCR and MLPA share the advantage of using uncultured cells versus karyotyping using cultured cells, in which selective pressure generally leads to lower grade chromosomal mosaicism, except for some rare cases of tissue-specific mosaicism.24 QF-PCR has been reported to detect mosaicism as low as 15%.7 It has been reported that MLPA is not expected to detect chromosomal mosaicism and that this still needs to be determined.10, 17 In our series, we found 9 of 4000 (0.23%) cases of aneuploidy mosaicism of which 7 were detected by MLPA. We have established cutoff levels for each probe to be able to discern chromosomal mosaicism. The rare case of trisomy 18 mosaicism in uncultured AF cells (as detected by MLPA and FISH), whereas cell cultures revealed a full-blown trisomy 18 may be attributed to tissue-specific mosaicism.24 In the case where MLPA and QF-PCR showed XYY/XY mosaicism, the cell cultures revealed 45,X[2]/46,XY[12] mosaicism. This could be explained by non-disjunction of a normal XY cell during early embryonic development with selection against the XYY cell line in AF cell cultures. If chromosomal mosaicism is suspected by visual inspection of the probe signals of the chromosomes involved, we advise to use additional genotyping such as FISH (for further determination of the level of mosaicism), as detection levels of 5% or more can be achieved as mentioned before.
The MLPA P095 kit is designed for rapid aneuploidy testing of a selection of chromosomes. The methodology, however, involves relative quantification of single probes in genes of known or unknown function. Theoretically, a deletion or an amplification of one or more probes could be the result of a structural (submicroscopic) chromosome anomaly. We have chosen to determine the sensitivity of each probe in detecting a (submicroscopic) chromosome anomaly. We have encountered 3 of 4000 (0.08%) cases with a structural chromosome defect initially found by an amplification and a deletion of one and two MLPA probe(s), respectively. An amplification of one probe on chromosome 13 was the result of a familial insertion of chromosome 13q12.3–q14.1 in chromosome 17. The deletion of two chromosome 21 probes could be explained by an unbalanced translocation of an inverted chromosome 4 and a chromosome 21. In addition, a submicroscopic deletion of the probe in the TYMS gene on chromosome 18p11.32 has been detected by interpreting single MLPA probe profiles. The indication for chromosome analysis was fetal anomalies on US examination. This deletion was confirmed by FISH with an overlapping probe (BAC).
This raises the question whether or not one should interpret the relative quantity of single probes in single genes. On the one hand, it has the obvious advantage of detecting a selection of structural chromosome defects,1, 7 but on the other hand it can also detect submicroscopic copy number variations from which the clinical significance is not known, like, for instance, in the TYMS gene, or ethically disputed like in the BRCA2 gene. This obviously leads to uncertainties in genetic counselling due to lack of knowledge of prenatal genotype–phenotype correlations as seen and anticipated with genomic microarrays. In addition, the interpretation of single probes in genes with known genotype–phenotype correlations, such as Duchenne's muscular dystrophy, would be indirect screening for a selection of single gene disorders. This can only be performed if certain criteria are met, which is not the case and is beyond the scope of common aneuploidy screening. Therefore, if MLPA is used as a stand-alone test, we advise to neglect the interpretation of the quantification of single probes to be able to screen only for microscopic-visible chromosome defects, which can be found by successive karyotyping.
We proved that MLPA is a reliable technique for RAD. As MLPA for prenatal diagnosis of common aneuploidies is now truly validated, the question raises whether MLPA can replace FISH and karyotyping for the most common indications.25, 26 For detection of the most common aneuploidies, FISH can be replaced by MLPA in most cases. MLPA is cheaper and less labor intensive compared with FISH, especially if more than five samples have to be processed. If karyotyping would also be replaced, some chromosome aberrations with a considerable risk of clinically significant congenital malformations/mental retardation would not be detected. The prevalence of these undetected cases is about 0.4% by fetal karyotyping and an estimated 0.1% in live borns.12, 25, 26 Currently, various studies show differences concerning the nature of chromosome aberrations considered to result in live born children with clinically significant abnormal phenotypes.25, 26, 27, 28 In our opinion, it is important to define which chromosome anomalies can be considered to be clinically significant and to confirm given estimates (manuscript in preparation). These are needed to determine how MLPA or QF-PCR can be implemented in prenatal diagnosis.
The most obvious indications to use MLPA as a stand-alone test are advanced maternal age and an enhanced risk of fetal Down's syndrome after prenatal screening. The individual risk determination after prenatal screening for Down's syndrome is expected to lead to more anxiety than the general determined risk on the basis of advanced maternal age. The latter indication group is declining after the introduction of prenatal screening. This requires large-scale RAD and therefore the introduction of MLPA for this might be a logical step forward.
Acknowledgments
We thank Stefanie van Veen and all other technicians of the Prenatal Cytogenetic Laboratory for their important contribution to this study. We also thank Dr DJJ Halley and E Wauters of the DNA diagnosis laboratory and all participating gynecologists for their contribution. We thank AJH Hamers for critically reviewing this manuscript and Kay Ballantyne for her contribution.
References
- Hultén MA, Dhanjal S, Pertl B. Rapid and simple prenatal diagnosis of common chromosome disorders: advantages and disadvantages of the molecular methods FISH and QF-PCR. Reproduction. 2003;126:279–297. doi: 10.1530/rep.0.1260279. [DOI] [PubMed] [Google Scholar]
- Winsor EJ, Silver MP, Theve R, Wright M, Ward BE. Maternal cell contamination in uncultured amniotic fluid. Prenat Diagn. 1996;16:49–54. doi: 10.1002/(SICI)1097-0223(199601)16:1<49::AID-PD808>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Witters I, Devriendt K, Legius E, et al. Rapid prenatal diagnosis of trisomy 21 in 5049 consecutive uncultured amniotic fluid samples by fluorescence in situ hybridisation (FISH) Prenat Diagn. 2002;22:29–33. doi: 10.1002/pd.225. [DOI] [PubMed] [Google Scholar]
- Stojilkovic-Mikic T, Mann K, Docherty Z, Mackie Ogilvie C. Maternal cell contamination of prenatal samples assessed by QF-PCR genotyping. Prenat Diagn. 2005;25:79–83. doi: 10.1002/pd.1089. [DOI] [PubMed] [Google Scholar]
- Donaghue C, Roberts A, Mann K, Ogilvie CM. Development and targeted application of a rapid QF-PCR test for sex chromosome imbalance. Prenat Diagn. 2003;23:201–210. doi: 10.1002/pd.569. [DOI] [PubMed] [Google Scholar]
- Cirigliano V, Voglino G, Canadas MP, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR. Assessment on 18 000 consecutive clinical samples. Mol Hum Reprod. 2004;10:839–846. doi: 10.1093/molehr/gah108. [DOI] [PubMed] [Google Scholar]
- Mann K, Donaghue C, Fox SP, Docherty Z, Ogilvie CM. Strategies for the rapid prenatal diagnosis of chromosome aneuploidy. Eur J Hum Genet. 2004;12:907–915. doi: 10.1038/sj.ejhg.5201224. [DOI] [PubMed] [Google Scholar]
- Grimshaw GM, Szczepura A, Hulten M, et al. Evaluation of molecular tests for prenatal diagnosis of chromosome abnormalities. Health Technol Assess. 2003;7:1–146. doi: 10.3310/hta7100. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes T, Kirchhoff M, Lind AM, Larsen GV, Schwartz M, Lundsteen C. Computer-assisted prenatal aneuploidy screening for chromosome 13, 18, 21, X and Y based on multiplex ligation-dependent probe amplification (MLPA) Eur J Hum Genet. 2005;13:171–175. doi: 10.1038/sj.ejhg.5201307. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Bui T-H. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am J Med Genet C Semin Med Genet. 2007;145:87–98. doi: 10.1002/ajmg.c.30114. [DOI] [PubMed] [Google Scholar]
- Bui T-H. Prenatal cytogenetic diagnosis: gone FISHing, BAC soon. Ultrasound Obstet Gynecol. 2007;30:247–251. doi: 10.1002/uog.5142. [DOI] [PubMed] [Google Scholar]
- Egberts J, Huisman M, van Leeuwen A, van Loon J. Improved method for determining fetal hemoglobin (HbF) by alkali denaturation. Clin Chem. 1995;41:1778–1780. [PubMed] [Google Scholar]
- Van Opstal D, Van Hemel JO, Sachs ES. Fetal aneuploidy diagnosed by fluorescence in-situ hybridisation within 24 hours after amniocentesis. Lancet. 1993;342:802. doi: 10.1016/0140-6736(93)91565-4. [DOI] [PubMed] [Google Scholar]
- Van Opstal D, van Hemel JO, Eussen BH, et al. A chromosome 21-specific cosmid cocktail for the detection of chromosome 21 aberrations in interphase nuclei. Prenat Diagn. 1995;15:705–711. doi: 10.1002/pd.1970150805. [DOI] [PubMed] [Google Scholar]
- Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, Choo KH. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA) J Med Genet. 2003;40:907–912. doi: 10.1136/jmg.40.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach R, Meijer J, van de Brug J, Vossebeld-Hoff I, et al. Rapid detection of chromosomal aneuploidies in uncultured amniocytes by multiplex ligation-dependent probe amplification (MLPA) Prenat Diagn. 2005;25:1032–1039. doi: 10.1002/pd.1247. [DOI] [PubMed] [Google Scholar]
- De Vigan C, Baena N, Cariati E, Clementi M, Stoll C. Contribution of ultrasonographic examination to the prenatal detection of chromosomal abnormalities in 19 centres across Europe. Ann Genet. 2001;44:209–217. doi: 10.1016/s0003-3995(01)01091-7. [DOI] [PubMed] [Google Scholar]
- Benn PA, Hsu LY. Maternal cell contamination of amniotic fluid cell cultures: results of a US nationwide survey. Am J Med Genet. 1983;15:297–305. doi: 10.1002/ajmg.1320150213. [DOI] [PubMed] [Google Scholar]
- Hockstein S, Chen PX, Thangavelu M, Pergament E. Factors associated with maternal cell contamination in amniocentesis samples as evaluated by fluorescent in situ hybridization. Obstet Gynecol. 1998;92 4 Part 1:551–556. doi: 10.1016/s0029-7844(98)00262-2. [DOI] [PubMed] [Google Scholar]
- Steinberg S, Katsanis S, Moser A, Cutting G. Biochemical analysis of cultured chorionic villi for the prenatal diagnosis of peroxisomal disorders: biochemical thresholds and molecular sensitivity for maternal cell contamination detection. J Med Genet. 2005;42:38–44. doi: 10.1136/jmg.2004.023556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V, Gesny R, Morichon-Delvallez N, et al. Prenatal diagnosis and normal outcome of a 46,XX/46,XY chimera: a case report. Hum Reprod. 2007;22:1037–1041. doi: 10.1093/humrep/del480. [DOI] [PubMed] [Google Scholar]
- Hook EB. Exclusion of chromosomal mosaicism: tables of 90%, 95% and 99% confidence limits and comments on use. Am J Hum Genet. 1977;29:94–97. [PMC free article] [PubMed] [Google Scholar]
- Van Opstal D, van den Berg C, Galjaard RJ, Los FJ. Follow-up investigations in uncultured amniotic fluid cells after uncertain cytogenetic results. Prenat Diagn. 2001;21:75–80. doi: 10.1002/1097-0223(200102)21:2<75::aid-pd990>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- Caine A, Maltby AE, Parkin CA, Waters JJ, Crolla JA. Prenatal detection of Down's syndrome by rapid aneuploidy testing for chromosomes 13, 18, and 21 by FISH or PCR without a full karyotype: a cytogenetic risk assessment. Lancet. 2005;366:123–128. doi: 10.1016/S0140-6736(05)66790-6. [DOI] [PubMed] [Google Scholar]
- Leung WC, Lao TT. Rapid aneuploidy testing, traditional karyotyping, or both. Lancet. 2005;366:97–98. doi: 10.1016/S0140-6736(05)66791-8. [DOI] [PubMed] [Google Scholar]
- Ogilvie CM, Lashwood A, Chitty L, Waters JJ, Scriven PN, Flinter F. The future of prenatal diagnosis: rapid testing or full karyotype? An audit of chromosome abnormalities and pregnancy outcomes for women referred for Down's syndrome testing. BJOG. 2005;112:1369–1375. doi: 10.1111/j.1471-0528.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Kagan KO, Molina FS, Waters JJ, Nicolaides KH. Fetal nuchal translucency scan and early prenatal diagnosis of chromosomal abnormalities by rapid aneuploidy screening: observational study. BMJ. 2006;332:452–455. doi: 10.1136/bmj.38730.655197.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]