Abstract
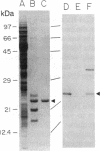
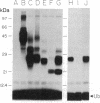
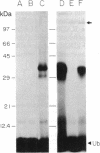
The RAD6 gene from the yeast Saccharomyces cerevisiae encodes a ubiquitin carrier protein (E2) required for a variety of cellular processes including DNA repair, induced mutagenesis, and sporulation. Here we identify an E2 from a higher plant, wheat, that is similar to RAD6 with respect to both structure and in vitro substrate specificity. The protein was purified from wheat germ by a combination of ubiquitin covalent affinity chromatography and anion-exchange HPLC and has an apparent molecular mass of 23 kDa [referred to as E2(23 kDa)]. E2(23 kDa) was capable of binding ubiquitin by means of a thiol ester linkage in an ATP-dependent and ubiquitin-activating enzyme-dependent reaction. In the presence of a variety of target proteins, E2(23 kDa), like the RAD6 gene product, formed covalent ubiquitin-protein conjugates in vitro only with histones in a ubiquitin protein ligase-independent reaction. E2(23 kDa) recognized both core and linker histones with an apparent order of preference of H2A greater than or equal to H1 greater than H2B greater than H3 greater than H4. This E2 protein was approximately 17-fold more effective at conjugating ubiquitin to histones than three other purified wheat germ E2 proteins tested. Mouse anti-E2(23 kDa) antibodies were used to isolate E2(23 kDa) DNA sequences from a wheat cDNA expression library. Antibody-positive clones were confirmed by amino acid identity of the sequence deduced from the cDNA to the peptide sequence of an E2(23 kDa) tryptic fragment. Protein expressed in Escherichia coli by the E2(23 kDa) cDNA was capable of both thiol ester adduct formation and conjugation of ubiquitin to histones. Analysis of the E2(23 kDa) cDNA shows that it encodes a protein with considerable amino acid sequence similarity to the yeast RAD6 gene product. Similarities exist at the amino terminus, the region surrounding the putative ubiquitin binding site, and at the carboxyl terminus, which is unusually acidic. Based on both the structural and enzymatic similarities to the RAD6 gene product, E2(23 kDa) may represent the first DNA repair enzyme identified in higher plants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chin D. T., Kuehl L., Rechsteiner M. Conjugation of ubiquitin to denatured hemoglobin is proportional to the rate of hemoglobin degradation in HeLa cells. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5857–5861. doi: 10.1073/pnas.79.19.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Elias S., Heller H., Hershko A. "Covalent affinity" purification of ubiquitin-activating enzyme. J Biol Chem. 1982 Mar 10;257(5):2537–2542. [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Katz-Etzion R., Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci U S A. 1981 Feb;78(2):761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DNA damage and repair in higher plants and their relation to genetic damage. Mutat Res. 1987 Nov;181(1):1–214. [PubMed] [Google Scholar]
- Dean C., Tamaki S., Dunsmuir P., Favreau M., Katayama C., Dooner H., Bedbrook J. mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 1986 Mar 11;14(5):2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988 Sep 9;241(4871):1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Goldknopf I. L., Wilson G., Ballal N. R., Busch H. Chromatin conjugate protein A24 is cleaved and ubiquitin is lost during chicken erythropoiesis. J Biol Chem. 1980 Nov 25;255(22):10555–10558. [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Haas A. L., Bright P. M., Jackson V. E. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988 Sep 15;263(26):13268–13275. [PubMed] [Google Scholar]
- Haas A. L., Bright P. M. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J Biol Chem. 1988 Sep 15;263(26):13258–13267. [PubMed] [Google Scholar]
- Haas A. L., Rose I. A. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982 Sep 10;257(17):10329–10337. [PubMed] [Google Scholar]
- Haas A. L., Warms J. V., Hershko A., Rose I. A. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982 Mar 10;257(5):2543–2548. [PubMed] [Google Scholar]
- Haas A. L., Wilkinson K. D. The large scale purification of ubiquitin from human erythrocytes. Prep Biochem. 1985;15(1-2):49–60. doi: 10.1080/00327488508062433. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin pathway for the degradation of intracellular proteins. Prog Nucleic Acid Res Mol Biol. 1986;33:19-56, 301. doi: 10.1016/s0079-6603(08)60019-7. [DOI] [PubMed] [Google Scholar]
- Hershko A., Eytan E., Ciechanover A., Haas A. L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982 Dec 10;257(23):13964–13970. [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983 Jul 10;258(13):8206–8214. [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Klemperer N. S., Berleth E. S., Pickart C. M. A novel, arsenite-sensitive E2 of the ubiquitin pathway: purification and properties. Biochemistry. 1989 Jul 11;28(14):6035–6041. doi: 10.1021/bi00440a047. [DOI] [PubMed] [Google Scholar]
- Matsui S. I., Seon B. K., Sandberg A. A. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Rose I. A. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985 Feb 10;260(3):1573–1581. [PubMed] [Google Scholar]
- Pickart C. M., Vella A. T. Ubiquitin carrier protein-catalyzed ubiquitin transfer to histones. Mechanism and specificity. J Biol Chem. 1988 Oct 15;263(29):15076–15082. [PubMed] [Google Scholar]
- Rechsteiner M. Ubiquitin-mediated pathways for intracellular proteolysis. Annu Rev Cell Biol. 1987;3:1–30. doi: 10.1146/annurev.cb.03.110187.000245. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Ubiquitin genes as a paradigm of concerted evolution of tandem repeats. J Mol Evol. 1987;25(1):58–64. doi: 10.1007/BF02100041. [DOI] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker S. Histone variants in plants. Evidence for primary structure variants differing in molecular weight. J Biol Chem. 1982 Dec 10;257(23):14250–14255. [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988 Nov;2(11):1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- Swerdlow P. S., Finley D., Varshavsky A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal Biochem. 1986 Jul;156(1):147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., Langan S. M., Haas A. L. Purification and initial characterization of ubiquitin from the higher plant, Avena sativa. J Biol Chem. 1985 Oct 5;260(22):12015–12021. [PubMed] [Google Scholar]
- Wu R. S., Kohn K. W., Bonner W. M. Metabolism of ubiquitinated histones. J Biol Chem. 1981 Jun 10;256(11):5916–5920. [PubMed] [Google Scholar]