Abstract
Mono-allelic germline mutations in mismatch repair (MMR) genes lead to Lynch syndrome, an autosomal dominant syndrome with an increased risk of predominantly colorectal and endometrial cancers. Bi-allelic germline mutations in MMR genes predispose to haematological malignancies, brain tumours, gastrointestinal tumours, polyposis and features of neurofibromatosis type 1 in early childhood.
We report a brother and a sister with bi-allelic germline mutations in MSH2; a pathogenic deletion of the first 6 exons and a variant of the initiation codon (c.1A>G (p.Met1?)), whereas their phenotypes (four colorectal cancers, small bowel carcinoma and 15 adenomas at age 39 and 48, and colorectal cancer, endometrial cancer and four adenomas at age 33 and 44, respectively) are more suggestive of a mono-allelic pathogenic MMR gene mutation. The carcinomas showed microsatellite instability in the presence of MLH1, PMS2, MSH2 and MSH6 proteins, indicating that the variant c.1A>G leads to an alternative protein with reduced activity that is retained in the tumours.
Our data suggest that the MSH2 variant c.1A>G (p.Met1?) should not be considered as a regular pathogenic mutation that leads to a strongly increased cancer risk, though it possibly contributes to a more severe phenotype when combined with a truncating mutation on the other allele.
Keywords: compound heterozygosity MSH2, mismatch repair gene variant, colorectal cancer, initiation codon
Introduction
Heterozygous germline mutations in mismatch repair (MMR) genes such as MLH1, MSH2, MSH6 and PMS2 lead to Lynch syndrome (or hereditary nonpolyposis colorectal cancer, HNPCC), a well-defined autosomal dominant cancer susceptibility syndrome with high penetrance. Mutation carriers are at high risk of developing colorectal and endometrial cancer and to a lesser extent cancer of small bowel, stomach, ovary, ureter and hepatobiliary tract at an average age of approximately 45 years.1 A hallmark of malignancies because of Lynch syndrome is contraction/expansion of repetitive sequences in tumour DNA, known as microsatellite instability (MSI).
Bi-allelic pathogenic germline mutations in MMR genes predispose to haematological malignancies, brain tumours and gastrointestinal tumours in early childhood and features of neurofibromatosis type 1 (NF1), especially café au lait patches (for review, see Felton et al2). Patients with pathogenic bi-allelic MMR gene mutations show microsatellite instability in gastrointestinal tumours.3, 4, 5, 6, 7, 8, 9, 10 In contrast, microsatellite stability has been observed in central nervous system tumours of patients with bi-allelic MMR gene mutations, suggesting a different pathway to tumorigenesis.2, 3 The clinical signs of NF1 in carriers of bi-allelic MMR gene mutations are attributed to somatic NF1 gene inactivation caused by defective MMR.11 Disruption of the NF1 gene might contribute to development of malignancies. In accordance with the phenotype of MMR gene knockout mouse models,12 all reported patients with pathogenic bi-allelic mutations in MLH1 and MSH2 developed malignancies. Bi-allelic mutations with residual protein function may lead to development of brain, haematological or gastrointestinal cancers in the second to fourth decades of life.2, 13
Interpretation of the consequences of mutations in MMR genes can be difficult. Some mutations, encoding truncated proteins, are clearly pathogenic, but the consequences of missense and silent mutations are less clear. We report two relatives with Lynch syndrome-associated malignancies developed in the fourth and fifth decade that carry bi-allelic mutations in the MSH2 gene; a pathogenic deletion of the first 6 exons (c.1–1076+?del) and a variant involving the initiation codon (c.1A>G (p.Met1?)). Observations in these relatives indicate that the altered initiation codon leads to a variant MSH2 protein, which probably has reduced, although not completely abolished, activity.
Materials and methods
Family data
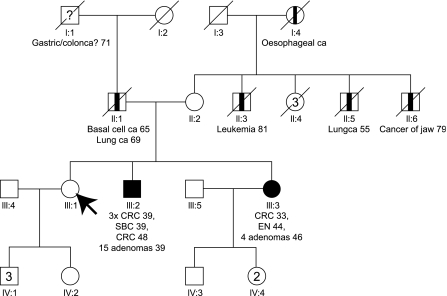
Initially a 45-year-old healthy female (III:1) was referred to the Department of Human Genetics of the Radboud University Nijmegen Medical Centre for genetic counselling because of the occurrence of colorectal cancer in her younger brother and sister. The pedigree is shown in Figure 1. Personal and family cancer histories were obtained. The medical history of the brother (III:2), sister (III:3) and father (II:1) was confirmed by reviewing pathological reports.
Figure 1.
Pedigree. → Arrow indicating initial counselee; CRC, Colorectal cancer; EN, Endometrial cancer; SBC, Small bowel cancer; ca, cancer; □ Male; ▪ Lynch syndrome-related cancer; ○ Female;  non-Lynch Syndrome-related cancer; / deceased; ? possible Lynch syndrome-related cancer (not verified).
non-Lynch Syndrome-related cancer; / deceased; ? possible Lynch syndrome-related cancer (not verified).
Individual III:2, the brother
The brother developed three synchronous colorectal carcinomas (in the caecum, ascending colon and an unspecified location) and synchronous cancer in the ileum (T3N1M0 stage IIIB) at age 39. Fifteen synchronous adenomas were found of which some showed high-grade dysplasia. A subtotal colectomy and partial resection of the ileum was performed. No features of neurofibromatosis type 1 were present. Colonic surveillance was offered but declined by the patient, as was physical examination. He developed a fourth primary colorectal cancer at age 48.
Individual III:3, the sister
The sister developed a colorectal cancer at age 33 in her caecum, T3N2M0 stage IIIC. A right-sided hemicolectomy was performed. An endometrioid type endometrial carcinoma without lymph node metastasis was diagnosed at age 44, treated with radical surgery. Physical examination and periodic colonoscopies took place every 1–2 years. At age 46, four adenomas with low-grade dysplasia were found in the colon, two proximally and two distally located. Physical examination of the sister (III:3) showed no signs of neurofibromatosis type I, except for a large head circumference (60 cm, +2.5 SD). Dermatological examination revealed several lenticular maculae diagnosed as naevi without signs of café au lait spots, freckling or neurofibromas.
Family history of cancer
The father had an excision of pre-auricular basal cell carcinoma at age 65. He died of poorly differentiated non-small cell lung carcinoma at age 69. In his family several relatives (paternal father, mother, uncle, aunt) possibly died of cancer, none of them could be verified.
The mother is, besides orthopaedic disabilities, healthy at age 80. Several of her relatives died of non-Lynch syndrome-associated cancer (three brothers and the mother).
Molecular studies
MSI analysis was performed in duplicate in four colorectal, one small bowel and one endometrial tumour using formalin-fixed paraffin-embedded tissue essentially as described by Hoogerbrugge et al.14 Areas with 60–80% tumour cells were used for analysis. The Bethesda panel of microsatellite markers15 was used together with the additional markers D1S158, D9S63, D18S58, D18S61, D8S199 and BAT40.
Immunohistochemistry was performed using antibodies against MLH1 (Pharmingen code: 51–1327gr), PMS2 (Pharmingen code: 556415), MSH2 (Oncogene Research Products code: NA26) and MSH6 (Transduction Laboratories code: G70220).
DNA extracted from peripheral blood lymphocytes of the sister (pedigree no III:3) was analysed for MLH1, MSH2 and MSH6 mutations by a combination of denaturing gradient gel electrophoresis, sequence analysis and multiplex ligation-dependent probe amplification (MLPA). Mutation analysis in the initial counselee (pedigree no III:1), brother (III:2) and mother (II:2) was performed by sequence analysis of exon 1 and MLPA of the MSH2 gene.
Results
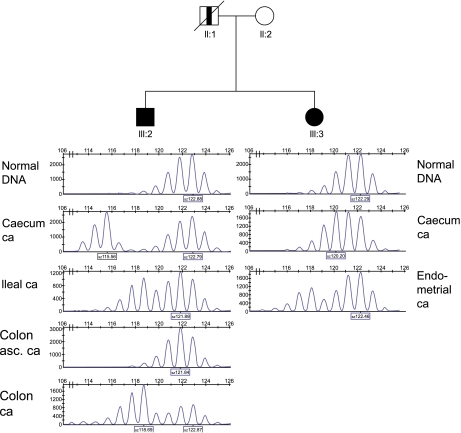
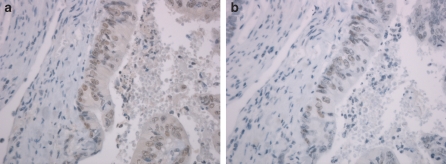
Lynch syndrome was suspected, because the pedigree (Figure 1) revealed the occurrence of early-onset and multiple Lynch syndrome-associated cancers in two first-degree relatives. Initially MSI analysis and immunohistochemical staining of the MMR proteins was performed on cancer of the caecum of the sister (III:3). The results of the MSI analysis with the Bethesda markers15 were inconclusive. Only the marker D2S123 showed subtle instability. The marker BAT25 did not show additional peaks in tumour DNA and was considered stable, although the pattern was shifted 2 bases to the left (Table 1, Figure 2). More markers were analysed which resulted in overall judgment that the colon tumour was MSI high, but with an overall microsatellite instability pattern different from instability patterns usually seen in tumours of patients with mismatch repair germline mutations. The immunohistochemical analysis of her colorectal tumour showed staining of MLH1, PMS2, MSH2 as well as MSH6. Because of the MSI high tumour without immunohistochemistry results indicating which gene to analyse first, mutation analysis of MLH1, MSH2 and MSH6 was performed in the sister (III:3). A pathogenic deletion of the first 6 exons (c.1-?_1076+?del) and a mutation of the initiation codon (c.1A>G (p.Met1?)) in exon one of the MSH2 gene was found. MSH2 mutation analysis in relatives showed presence of both mutations in the brother (III:2), absence of both mutations in the initially referred counselee (III:1) and presence of the c.1A>G-allele in the healthy mother. To examine whether instability was present in more tumours developed by the brother (III:2) and sister (III:3) and to gain insight in the carcinogenesis of these tumours, five additional tumours were analysed for MSI and immunohistochemistry. All tested tumours of the sister and the brother were MSI high (Table 1). In the six tumours tested 9 out of 18 (50%) BAT marker results and 40 out of 45 (89%) interpretable dinucleotide results were scored as obviously instable. Immunohistochemical staining of the MSH2 protein was weak and the concentration of the antibodies was increased to improve the results. No difference in staining intensity was observed between normal cells and tumour cells. MSH2 (Figure 3a), MLH1 and PMS2 protein staining was observed in all tumours. MSH6 protein (Figure 3b) was observed in 5/6 tumours, whereas judgment of the immunohistochemical pattern was inconclusive in 1/6 MSH6 stainings. In addition, staining of all four MMR proteins was found by immunohistochemical analysis in three low-grade adenomas of the sister (III:3).
Table 1. MSI and immunohistochemical analyses of lesions of subjects III:2 and III:3.
| MSI patterns of tested microsatellite markers | Immunohistochemistry | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tested material | D2S123 | D5S346 | D17S250 | BAT25 | BAT26 | BAT40 | D1S158 | D9S63 | D18S58 | D18S61 | D8S199 | MLH1 | PMS2 | MSH2 | MSH6 |
| Caecum ca III:3 | + | s | s | s-2 | s | s-1 | ? | + | + | + | + | p | p | p | p |
| Endometrial ca III:3 | + | + | + | + | s | + | + | + | + | + | ? | p | p | p | p |
| Adenoma III:3 | p | p | p | p | |||||||||||
| Caecum ca III:2 | + | + | + | + | s | + | + | - | + | + | + | p | p | p | x |
| Ileal carcinoma III:2 | + | + | + | + | + | + | + | + | + | + | + | p | p | p | p |
| Colon asc ca III:2 | ? | s | + | s-1 | s | s-2 | + | + | + | + | + | p | p | p | p |
| Colon ca III:2 | + | s | + | + | s | + | + | + | + | + | + | p | p | p | p |
+, instability; s, stable; ?, addition of 1 dinucleotide or loss of heterozygosity; x, inconclusive; s-1, peak shift of one basepair (considered stable); s-2, peak shift of two basepairs (considered stable); ca, cancer; asc, ascendens; p, present staining.
Figure 2.
BAT25 MSI patterns of tumours of individuals III:2 and III:3. One and two bp shift is observed in colon ascendens cancer of individual III:2 and caecum cancer of individual III:3, respectively. Asc, ascendens; Ca, cancer. □ Male; ▪ Lynch syndrome-related cancer; ○ Female;  non-Lynch syndrome related cancer; / deceased.
non-Lynch syndrome related cancer; / deceased.
Figure 3.
Examples of immunohistochemistry of MSH2 and MSH6 in caecum cancer of individual III:3. Presence of MSH2 (a) an MSH6 (b) staining in caecum cancer of individual III:3.
Discussion
The weak MSH2 staining in both normal and tumour tissues of both our patients indicates that the allele with variant c.1A>G generates a product which can be recognized by the antibody against MSH2. Most likely an MSH2 protein with a different N terminus is produced because of the use of a second in frame initiation codon, 26 codons downstream of the normally used initiation codon. The wild type initiation codon has a strong Kozak sequence with a G at position −3 and a G at position +4. The Kozak sequence of the alternative initiation codon is less strong but still adequate with a G at position −3 and a C at position +4.16 In the ileal carcinoma of III:2 instability of BAT26 is observed. As BAT26 is located in intron 5 of the MSH2 gene and, thus, is not present in the allele with the deletion of the first 6 exons, this indicates the c.1A>G-allele is present in that tumour. Moreover the preservation of the weak MSH2 protein expression in tumour cells of all six tumours tested in this family also suggests that no somatic inactivating mutation leading to complete protein loss of the c.1A>G-allele has occurred in these tumours, whereas in virtually all mismatch repair-deficient tumours of pathogenic MSH2 mutation carriers somatic mutations lead to loss of MSH2 and/or MSH6 protein expression.17, 18 Owing to the putative use of the second initiation codon, only a small region of the mismatch-recognition domain of MSH2 is missing, whereas the other functional domains of the protein remain intact.19 This might explain the residual but lower activity of the protein, which is reflected in the relatively subtle microsatellite instability in the tumours of our patients. Only 9/18 (50%) of the mononucleotide markers BAT25, BAT26 and BAT40 were obviously instable, whereas tumours of Lynch syndrome patients are known to show instability in more than 95% of mononucleotides.20, 21 Taken together, our data indicate that malfunction of the mismatch repair occurs without somatic inactivation of the c.1A>G-allele, which suggests reduced activity of the variant c.1A>G MSH2 protein. Neither of the affected siblings showed haematological malignancies, brain tumours or café au lait spots. Thus, compound heterozygosity for the pathogenic deletion of the first 6 exons and the variant c.1A>G of the MSH2 gene in the two individuals in this report is not associated with the phenotype that has been reported to be present in individuals with bi-allelic pathogenic MMR gene mutations.2 A deletion of the first 6 exons of MSH2 has been described previously and leads to a ‘classical Lynch syndrome' phenotype.22 The phenotype of the brother is more severe when compared with classical Lynch syndrome patients because he has multiple synchronous colorectal cancers and also metachronous cancer and more than 10 polyps. Lynch syndrome patients have a slightly increased risk of adenomas (but not more than 10) and have an increased risk of multiple tumours, but mostly two and not more at a time. The family history of the mother, carrier of the c.1A>G-variant, is not suggestive for Lynch syndrome. Both the absence of a bi-allelic pathogenic MMR gene mutation phenotype and lack of Lynch syndrome-associated carcinomas in the maternal family suggest that the effect of the c.1A>G-variant is not as severe as that of a truncating mutation. For that reason presymptomatic testing for the variant c.1A>G is not offered to relatives at the moment.
Heterozygosity for the pathogenic deletion is assumed in the deceased father. His family history is difficult to interpret as he is an only child. Paternal family history is suggestive of a predisposition of cancer, although this could not be verified.
Two other variants of the initiation codon of MSH2 have been described in two CRC patients (c.1A>T23 and c.1A>C24) and one undefined patient (c.1A>C25). Both mutations were considered pathogenic.23, 24 In carriers of these initiation codon variants the tumour was not MSI high and there was no family history of colorectal cancer. Moreover, immunohistochemical staining of MLH1, MSH2 and MSH6 was positive in the CRC of the carrier of variant c.1A>C.24 This is in line with our observation that the variant of the initiation codon of the MSH2 gene does not seem to lead to a classical Lynch syndrome phenotype. Functional studies are necessary to define the consequences of these variants.
The family reported here shows that compound heterozygosity for the pathogenic deletion of the first 6 exons and the variant c.1A>G of the MSH2 gene is not associated with childhood tumours or major features of neurofibromatosis type 1. The variant c.1A>G results in a variant MSH2 protein, which most likely has residual activity. This demonstrates that mutations of initiation codons, that are generally considered to be pathogenic, should be interpreted with caution. Moreover it illustrates the strength of studying the co-occurrence of mutations for the interpretation of unclassified variants, which is becoming an increasingly important subject in clinical molecular genetics.
References
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Felton K, Gilchrist D, Andrew S. Constitutive deficiency in DNA mismatch repair. Clin Genet. 2007;71:483–498. doi: 10.1111/j.1399-0004.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Charbonnier F, Moerman A, et al. Early onset brain tumor and lymphoma in MSH2-deficient children. Am J Hum Genet. 2003;72:213–216. doi: 10.1086/345297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M, Fasano C, Panariello L, et al. Evidence for a recessive inheritance of Turcot's syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR, Liu B, Parsons RE, et al. The molecular basis of Turcot's syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- Ostergaard JR, Sunde L, Okkels H. Neurofibromatosis von Recklinghausen type I phenotype and early onset of cancers in siblings compound heterozygous for mutations in MSH6. Am J Med Genet A. 2005;139:96–105. doi: 10.1002/ajmg.a.30998. [DOI] [PubMed] [Google Scholar]
- Trimbath JD, Petersen GM, Erdman SH, Ferre M, Luce MC, Giardiello FM. Cafe-au-lait spots and early onset colorectal neoplasia: a variant of HNPCC. Fam Cancer. 2001;1:101–105. doi: 10.1023/a:1013881832014. [DOI] [PubMed] [Google Scholar]
- Whiteside D, McLeod R, Graham G, et al. A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe-au-lait spots. Cancer Res. 2002;62:359–362. [PubMed] [Google Scholar]
- Will O, Carvajal-Carmona LG, Gorman P, et al. Homozygous PMS2 deletion causes a severe colorectal cancer and multiple adenoma phenotype without extraintestinal cancer. Gastroenterology. 2007;132:527–530. doi: 10.1053/j.gastro.2006.11.043. [DOI] [PubMed] [Google Scholar]
- Wang Q, Montmain G, Ruano E, et al. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am J Med Genet C Semin Med Genet. 2004;129:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- Felton K, Gilchrist D, Andrew S. Constitutive deficiency in DNA mismatch repair: is it time for Lynch III. Clin Genet. 2007;71:499–500. doi: 10.1111/j.1399-0004.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- Hoogerbrugge N, Willems R, Van Krieken HJ, et al. Very low incidence of microsatellite instability in rectal cancers from families at risk for HNPCC. Clin Genet. 2003;63:64–70. doi: 10.1034/j.1399-0004.2003.630110.x. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Mangold E, Pagenstecher C, Friedl W, et al. Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining. J Pathol. 2005;207:385–395. doi: 10.1002/path.1858. [DOI] [PubMed] [Google Scholar]
- Overbeek LI, Kets CM, Hebeda KM, et al. Patients with an unexplained microsatellite instable tumour have a low risk of familial cancer. Br J Cancer. 2007;96:1605–1612. doi: 10.1038/sj.bjc.6603754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de WN, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Kets CM, van Krieken JH, Hebeda KM, et al. Very low prevalence of germline MSH6 mutations in hereditary non-polyposis colorectal cancer suspected patients with colorectal cancer without microsatellite instability. Br J Cancer. 2006;95:1678–1682. doi: 10.1038/sj.bjc.6603478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R. Quasimonomorphic mononucleotide repeats for high-level microsatellite instability analysis. Dis Markers. 2004;20:251–257. doi: 10.1155/2004/159347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Barrows A, Wijnen JT, et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet. 2003;72:1088–1100. doi: 10.1086/373963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington SM, Lin-Goerke J, Ling J, et al. Systematic analysis of hMSH2 and hMLH1 in young colon cancer patients and controls. Am J Hum Genet. 1998;63:749–759. doi: 10.1086/301996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- Otway R, Tetlow N, Hornby J, Kohonen-Corish M. Evaluation of enzymatic mutation detection trade mark in hereditary nonpolyposis colorectal cancer. Hum Mutat. 2000;16:61–67. doi: 10.1002/1098-1004(200007)16:1<61::AID-HUMU11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]