Abstract
An imbalance between excitation and inhibition in the cerebral cortex has been suggested as a possible etiology of autism. The DLX genes encode homeodomain-containing transcription factors controlling the generation of GABAergic cortical interneurons. The DLX1 and DLX2 genes lie head-to-head in 2q32, a region associated with autism susceptibility. We investigated 6 Tag SNPs within the DLX1/2 genes in two cohorts of multiplex (MPX) and one of simplex (SPX) families for association with autism. Family-based association tests showed strong association with five of the SNPs. The common alleles of rs743605 and rs4519482 were significantly associated with autism (P<0.012) in the first sample of 138 MPX families, with the latter remaining significant after correction for multiple testing (Pcor=0.0046). Findings in a second sample of 169 MPX families not only confirmed the association at rs4519482 (P=0.034) but also showed strong allelic association of the common alleles at rs788172, rs788173 and rs813720 (Pcor=0.0003–0.04). In the combined MPX families, the common alleles were all significantly associated with autism (Pcor=0.0005–0.016). The GGGTG haplotype was over transmitted in the two MPX cohorts and the combined samples [Pcor<0.05: Pcor=0.00007 for the combined MPX families, Odds Ratio: 1.75 (95% CI: 1.33–2.30)]. Further testing in 306 SPX families replicated the association at rs4519482 (P=0.033) and the over transmission of the haplotype GGGTG (P=0.012) although P-values were not significant after correction for multiple testing. The findings support the presence of two functional polymorphisms, one in or near each of the DLX genes that increase susceptibility to, or cause, autism in MPX families where there is a greater genetic component for these conditions.
Keywords: autism spectrum disorders, epilepsy, genetics, DLX1 and DLX2 genes, GABAergic system, gene association
Introduction
Autism Spectrum Disorders (ASDs) are a group of neurodevelopmental disorders known as pervasive developmental disorders, characterized by impairments in social interaction and communication, and the presence of restricted activities and interests. ASDs are life-long conditions that are heterogeneous in etiology and highly variable in phenotype. Genetic epidemiological studies (twin,1, 2, 3 sibling4, 5 and family6, 7 studies) have provided considerable evidence for the presence of genetic factors in autism. ASDs have been proposed to result from the interaction of multiple genes and the interaction of genes and environmental factors.
Based in part on the high prevalence of epilepsy and seizures in individuals with autism,8 it has recently been suggested that an imbalance in the ratio of excitatory to inhibitory impulses in the brain may contribute to the development of autism by causing hyperexcitability in the cortex.9, 10 Rubenstein and Merzenich11 proposed an elegant model for autism in which there is an increased ratio of excitation to inhibition in those neural circuits that are responsible for language development and social behavior. They support their hypothesis with an extensive review of studies focused on understanding neural systems that are involved in memory, affiliative behaviors, and sensory–perceptual processing, and the modulation and integration of these systems, all of which are compromised or abnormal in individuals with autism. In their comprehensive review of the neuroanatomy and neurotransmitter systems in autism, Polleux and Lauder9 further suggest that an imbalance between excitation and inhibition in the cortex, resulting from genetic and environmental factors affecting either the pattern of neuronal populations and/or the synaptic assembly of these neuronal networks, provides a compelling model for the underlying neurobiological defects that result in autistic behaviors. Therefore, the investigation of genes that control the growth of populations of inhibitory or excitatory neurons as potential autism candidate genes is warranted. Among these, the DLX genes, which encode a family of homeobox transcription factors that are expressed during forebrain development,12 are attracting growing interest.
Mouse models have revealed the involvement of the Dlx genes in the differentiation and migration of GABAergic neurons. Both differentiation and migration of striatal interneurons are blocked in Dlx1/2-null mice,13, 14 and GABAergic neurons fail to migrate to the cerebral cortex.15, 16 Although Dlx1/2 and Dlx2 ablations are fatal, Dlx1-null mice are born healthy, but experience a time-dependent reduction in GABAergic interneurons in the hippocampus,17 a brain region involved in learning and memory. Post-mortem studies have implicated abnormalities in the development of the hippocampus in autism.9 In Dlx1-null mice, the reduction in GABAergic inhibitory impulses also causes late-onset epilepsy,17 which is more prevalent in individuals with autism than in the general population.8 Both Dlx2 and Dlx5 induce expression of glutamic acid decarboxylases,18 which are involved in the synthesis of GABA. A functional alteration in DLX genes may alter the differentiation and migration of GABAergic interneurons, consequently changing the inhibition/excitation ratio in some parts of the brain. Thus we have hypothesized that the DLX genes play a role in ASDs through their proven involvement in the development of the GABAergic system.
DLX genes are arranged in bi-gene clusters, two of which, the DLX1/DLX2 and DLX5/DLX6 clusters, are located in chromosomal regions 2q32 and 7q22, respectively. These regions have shown association with autism susceptibility in genome scan studies,19 making these DLX genes good positional candidates for ASDs.
Although the two previous association studies examining single-nucleotide polymorphisms (SNPs) in the DLX2 and DLX6 genes did not find association with autism,20, 21 these studies were limited to the analysis of the coding regions of the genes. The study by Hamilton et al22 of rare variants in the DLX1/DLX2 and DLX5-DLX6 regions in families with autistic children also did not establish an association of these variants with autism susceptibility; however, only those variants leading to non-synonymous amino acid substitutions were investigated. Therefore, we investigated 6 Tag SNPs in the region covering the DLX1 and DLX2 genes in two independently recruited cohorts of multiplex (MPX) families, as well as a simplex (SPX) cohort, for association with susceptibility to autism. Our findings on the two MPX cohorts strongly support a role for these genes in the etiology of ASDs.
Materials and methods
We performed the study in a two-stage design. Sample 1 consisted of DNA samples from 138 MPX families purchased from the Autism Genetics Resource Exchange (AGRE: www.agre.org), including 288 affected children (219 males and 69 females). A second group of families, Sample 2, consisted of 169 MPX families (208 affected males and 145 affected females) recruited from across North America by the Autism Spectrum Disorders – Canadian–American Research Consortium (ASD-CARC; www.AutismResearch.ca). In addition, we examined 306 SPX families recruited by ASD-CARC, including 256 families with affected males and 50 with affected females. All MPX families had at least two affected siblings. Family selection criteria for the Sample 1 families from AGRE have been described previously.23, 24 Diagnoses of the affected individuals varied in the AGRE families, with 220 meeting criteria for autism, 20 for ‘not quite autism' and 45 for ‘broad spectrum'. The designations ‘not quite autism' and ‘broad spectrum' are not DSM-IV diagnoses but are defined by AGRE (http://www.agre.org/agrecatalog/algorithm.cfm). The diagnoses of the affected individuals from the Sample 2 and SPX families were based on multiple approaches including comprehensive assessments by collaborating ASD-CARC clinical geneticist-led interdisciplinary teams and Autism Diagnostic Interview-Revised (ADI-R)25 and/or ADOS26 or PDD Behavior Inventory (PDDBI)27, 28 data. All of the affected individuals in the MPX families had a clinical standard diagnosis of ASD by no less than the ADI-R and ADOS and standardization according to DSM-IV criteria. Among the 306 affected children in the SPX families, 262 have ADI-R data available, all meeting diagnostic criteria for autism or not quite autism. PDDBI information was available for the remaining and all met the cutoff for autism or PDD.
This study was approved by the Research Ethics Board of all participating ASD-CARC institutions including Queen's University, New York State Institute for Basic Research in Developmental Disabilities, University of Manitoba, and University of British Columbia. Written informed consent was obtained from parents of all participating family members from Canada and the United States (and through AGRE for the AGRE families).
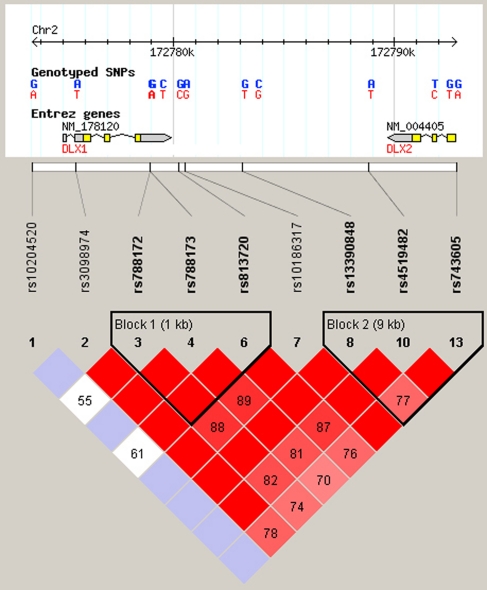
SNPs were selected based on haplotype structure information available from the HAPMAP project (http://www.hapmap.org/). Tag SNPs were identified with the assistance of the Haploview program (v3.32) (http://www.broad.mit.edu/mpg/haploview/)29 and the Applied Biosystems SNPbrowser™ software (v3.5) (http://www.appliedbiosystems.com), using the following criteria: (1) SNPs with minor allele frequencies of >0.05 and (2) CEPH samples as a reference group because our ASD families are mostly of European descent. Six Tag SNPs in a 30-kb flanking region of the DLX1 and DLX2 genes were selected for study (see Figure 1).
Figure 1.
Linkage disequilibrium (LD) map of the DLX1/DLX2 genes. Thirteen SNP markers within the 30 kb of genomic sequence spanning the DLX1/DLX2 genes. Pairwise LD among SNPs was estimated using D′ with Haploview. The number inside each rhombus is a D′ value*100. The structure and position of the DLX1/DLX2 genes, the positions of the 6 Tag SNPs for genotyping are indicated (SNP markers in bold).
SNP rs788172 was genotyped using traditional RFLPs. A 217-bp fragment covering this SNP was amplified by PCR using the following primers 5′-CTCTGTCTGTGCGCTGGTAA-3′ and 3′-AGGCGAAGTCCATTTCTCAA-5′. PCR was performed on a Peltier Thermal Cycler (MJ Technologies) using 8 ng DNA in a reaction volume of 4 μl, containing 1 × PCR buffer (Invitrogen), 4 × 0.2 m dNTP, 24 ng each of the forward and reverse primers, 2.0 m MgCl2, and 0.2 U Taq DNA Polymerase (Invitrogen). PCR consisted of 30 cycles of 45 s at 95°C, 30 s at 61°C, and 30 s at 72°C. Cycling was preceded by a 5 min denaturation step at 95°C and followed by a 6 min incubation at 72°C. PCR products were digested with PstI restriction endonuclease (New England Biolabs). Digestions were carried out overnight at 37°C in a reaction volume of 7 μl, containing 15 m MgCl2 (Invitrogen), 1 × PCR buffer, 2 × BSA (New England Biolabs), 0.67 m DTT, 0.6 U of PstI restriction enzyme, and 4 μl of PCR product. Electrophoresis of digested samples was carried out in a 2% agarose gel and the DNA fragments were stained with ethidium bromide. Samples were scored based on the presence of the expected bands for each allele and each result was read independently by two persons.
Genotyping of the other five SNPs was carried out using validated custom TaqMan SNP Genotyping Assays (http://www.appliedbiosystems.com) on an ABI Prism 7900HT, using 384-well plates. Duplicate samples and negative controls were included in each plate to check the accuracy of genotyping. Genotypes were automatically scored with the SDS 2.2.2 software using standard parameters.
Before data analysis, each polymorphism was assessed in parents and affected cases using χ2 for deviations from Hardy–Weinberg equilibrium with the HWE program (Ott J, 1999).30 This step was performed to determine whether there was any departure from HWE because such a finding can suggest that the marker is linked to a susceptibility or protective allele.
Genotypes were checked for Mendelian inconsistencies using the FBAT program (v1.7.3)31 and all identified Mendelian errors were either corrected by regenotyping individual samples or omitted from data analysis. Linkage disequilibrium (LD) of each pair of SNPs was assessed using the 2LD program.32 To perform family-based single-marker and multi-locus tests of association, the FBAT program (v1.7.3) was used.31 FBAT is an extension of the original transmission disequilibrium test.33 It uses a generalized score statistic to perform a variety of transmission disequilibrium tests for single marker alleles, haplotypes and genotypes in the presence or absence of linkage. Because FBAT uses conditional distributions in deriving the distribution for the test statistic under the null hypothesis, biases owing to population admixture, mis-specification of the trait distribution, and/or selection based on a trait can be avoided. FBAT uses data from related triads and therefore can handle MPX families when testing for transmission disequilibrium. All family-based association tests in this study were performed using an additive model under the null hypothesis of no association in the presence of linkage. The individual haplotype tests were conducted under the ‘biallelic' mode in haplotype FBAT.
Corrections for multiple comparisons
As the SNPs that we examined show a high degree of intermarker LD (ie, non-independence of the tests) and there are two haplotype sub-blocks, we corrected for multiple comparisons by applying a correction factor of 8 (2 (samples) × 2 (test-retest) × 2 (haplotype sub-blocks)) for all the tests. The corrected P-values (Pcor) were calculated by multiplying the P-values by eight. Results were considered significant at Pcor<0.05.
Estimating odds ratios
Odds Ratios were calculated according to the suggestions of Ackerman et al, 2005,34 by comparing the observed number of transmitted alleles or haplotypes and the number of non-transmitted alleles or haplotypes. The number of non-transmitted alleles or haplotypes were calculated by multiplying the number of expected transmissions by two and subtracting the number of observed transmissions.
Results
Figure 1 shows the locations of the six SNPs tested. The alleles in the cases and parents from both sets of MPX families and the SPX families were in Hardy–Weinberg Equilibrium (HWE).
The results of the single marker FBAT analyses are summarized in Table 1. In the single marker FBAT tests in Sample 1 families, we found highly significant preferential transmission of the common T allele of rs4519482 [P=0.0006, Pcor=0.0046, OR=1.96 (95% CI: 1.44–2.76)] and significant preferential transmission of the common G allele of rs743605 [P=0.012, OR=1.74 (95% CI: 1.26–2.41)] although this was marginal after correction (Pcor=0.096). Both SNPs are located in DLX2. The remaining four SNPs (rs788172, rs788173, rs813720 and rs13390848), which are in or near the DLX1 gene, did not show significant association although there was a tendency towards over transmission of the common alleles for three of the SNPs: rs788172-G, rs788173-G and rs813720-G.
Table 1. Single SNP FBAT for association of 6 Tag SNPs in the DLX1/DLX2 genes in MPX families with ASD.
| Sample 1 MPX (N=138) | Sample 2 MPX (N=169) | Combined MPX (N=307) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Allele | S | E(S) | Z | P | Pcor | S | E(S) | Z | P | Pcor | S | E(S) | Z | P | Pcor |
| rs788172 | A | 139 | 152 | −1.617 | 0.11 | 101 | 121 | −2.814 | 0.0049 | 0.039 | 240 | 273 | −3.095 | 0.002 | 0.016 | |
| G | 247 | 235 | 1.617 | 223 | 203 | 2.814 | 470 | 438 | 3.095 | |||||||
| rs788173 | A | 135 | 145 | −1.256 | 0.21 | 89 | 118 | −4.108 | 0.00004 | 0.0003 | 224 | 263 | −3.72 | 0.0002 | 0.0016 | |
| G | 239 | 230 | 1.256 | 229 | 200 | 4.108 | 468 | 430 | 3.72 | |||||||
| rs813720 | C | 141 | 155 | −1.725 | 0.085 | 107 | 133 | −3.4 | 0.0007 | 0.0054 | 249 | 288 | −3.548 | 0.0004 | 0.0031 | |
| G | 245 | 232 | 1.725 | 249 | 224 | 3.4 | 495 | 457 | 3.548 | |||||||
| rs13390848 | T | 106 | 103 | 0.494 | 0.62 | 116 | 100 | 2.388 | 0.017 | 0.14 | 222 | 202 | 2.02 | 0.043 | ||
| G | 218 | 222 | −0.494 | 190 | 207 | −2.388 | 408 | 428 | −2.02 | |||||||
| rs4519482 | A | 97 | 123 | −3.441 | 0.0006 | 0.0046 | 84 | 98 | −2.123 | 0.034 | 181 | 221 | −3.982 | 0.00007 | 0.0005 | |
| T | 217 | 192 | 3.441 | 166 | 152 | 2.123 | 383 | 344 | 3.982 | |||||||
| rs743605 | A | 139 | 159 | −2.5 | 0.012 | 0.096 | 171 | 181 | −1.24 | 0.21 | 310 | 340 | −2.641 | 0.0083 | 0.066 | |
| G | 213 | 193 | 2.5 | 231 | 221 | 1.24 | 444 | 414 | 2.641 | |||||||
Pcor: Corrected P-values. See Methods for a description of the correction applied for multiple testing.
The T allele of rs4519482 was significantly over transmitted in the Sample 2 families, although the significance was not as strong as in the Sample 1 families. The significant association of rs743605 in the Sample 1 families was not evident in the Sample 2 families. Interestingly, three common alleles at rs788172-G, rs788173-G and rs813720-G, which showed only a tendency towards over transmission in the Sample 1 families, were very significantly over transmitted from parents to affected offspring [Pcor=0.039, 0.0003 and 0.0054, respectively; OR=1.70 (95% CI: 1.23–2.35), 2.21 (95% CI: 1.59–3.08) and 1.86 (95% CI: 1.53–2.37), respectively]. Unlike in the Sample 1 families, the rs13390848-G allele was also over transmitted in the Sample 2 families [P=0.017, OR=1.63 (95% CI: 1.16–2.29)], although this was not significant after correction.
All of the common alleles of the markers were significantly over transmitted in the combined MPX families (Table 1).
We also tested the 6 Tag SNPs in 306 SPX families. Only the common T allele at rs4519482 showed an association with ASD [P=0.033, OR=1.59 (95% CI: 1.11–2.27)] before correction (Table 2).
Table 2. Single SNP FBAT for association of 6 Tag SNPs in DLX1/DLX2 in SPX families with ASD.
| SNP | Allele | S | E(S) | Z | P | Pcor |
|---|---|---|---|---|---|---|
| rs788172 | A | 137 | 143 | −0.715 | 0.47 | |
| G | 233 | 228 | 0.715 | |||
| rs788173 | A | 154 | 159 | −0.553 | 0.58 | |
| G | 256 | 252 | 0.553 | |||
| rs813720 | C | 162 | 169 | −0.784 | 0.43 | |
| G | 262 | 256 | 0.784 | |||
| rs13390848 | T | 109 | 104 | 0.712 | 0.48 | |
| G | 213 | 218 | −0.712 | |||
| rs4519482 | A | 85 | 99 | −2.16 | 0.033 | 0.26 |
| T | 173 | 159 | 2.16 | |||
| rs743605 | A | 186 | 187 | −0.122 | 0.9 | |
| G | 222 | 221 | 0.122 |
Pcor, Corrected P-values. See Materials and methods for a description of the correction applied for multiple testing.
As the common alleles at rs788172, rs788173, rs813720, rs4519482, and rs743605 were individually found to be associated with ASD, we next tested whether a haplotype consisting of these alleles was over transmitted in the different family groups. Strikingly, a haplotype composed of rs788172-G, rs788173-G, rs813720-G, rs4519482-T, and rs743605-G was significantly over transmitted in both sets of MPX families, as well as in the combined MPX families [Pcor=0.036, 0.0022 and 0.00007, respectively; OR=1.89 (95% CI: 1.25–2.86), 1.96 (95% CI: 1.33–2.9) and 1.75 (95% CI: 1.33–2.30) respectively] (Table 3). There was also a tendency towards over transmission of this haplotype in the SPX families (Pcor=0.096)
Table 3. Family-based haplotype association for the rs788172, rs788173, rs813720, rs4519482, and rs743605 SNPs in the DLX1/DLX2 genes in families with ASD.
| Haplotypes | Family set | Frequency | S | E(S) | Z | P | Pcor |
|---|---|---|---|---|---|---|---|
| Sample 1 MPX | 0.35 | 90 | 76 | 2.84 | 0.0045 | 0.036 | |
| GGGTG | Sample 2 MPX | 0.34 | 103 | 85 | 3.63 | 0.00028 | 0.0022 |
| Combined MPX | 0.34 | 193 | 163 | 4.44 | 0.00001 | 0.00007 | |
| SPX | 0.37 | 140 | 124 | 2.51 | 0.012 | 0.096 | |
| GGG- - | Sample 1 MPX | 0.637 | 229 | 214 | 1.93 | 0.054 | |
| Sample 2 MPX | 0.65 | 222 | 195 | 3.97 | 0.00007 | 0.00058 | |
| Combined MPX | 0.644 | 451 | 410 | 4.00 | 0.00006 | 0.0005 | |
| SPX | 0.64 | 214 | 208 | 0.73 | 0.47 | ||
| - - -TG | Sample 1 MPX | 0.384 | 116 | 98 | 3.14 | 0.0017 | 0.014 |
| Sample 2 MPX | 0.42 | 130 | 120 | 1.83 | 0.068 | ||
| Combined MPX | 0.405 | 246 | 218 | 3.54 | 0.00042 | 0.0034 | |
| SPX | 0.433 | 149 | 139 | 1.47 | 0.14 |
Pcor, Corrected P-values. See Materials and methods for a description of the correction applied for multiple testing.
Based on haplotype structure information from HAPMAP (Figure 1) and the data from this study (not shown), rs788172, rs788173, rs813720, rs4519482 and rs743605 are in the same major haplotype block. However, it is clear that there are two sub-blocks, with rs788172, rs788173 and rs813720 being in the first sub-block covering the DLX1 gene and rs4519482 and rs743605 in the sub-block covering the DLX2 gene. We therefore performed haplotype transmission tests on the two sub-blocks of haplotypes and, similar to the single marker analysis, we found: (1) the rs788172-G rs788173-G rs813720-G haplotype in the first sub-block was very significantly over transmitted in the Sample 2 and combined MPX families [Pcor=0.00058 and 0.0005, respectively; OR=1.88 (95% CI: 1.32–2.69) and 1.57 (95% CI: 1.25–1.97) respectively] (Table 3), with marginal and no significance in Sample 1 MPX families and SPX families, respectively; (2) in contrast, the rs4519482-T rs743605-G haplotype in the second sub-block showed highly significant preferential transmission in Sample 1 and combined MPX families [Pcor=0.014 and 0.0034; OR=1.85 (95% CI: 1.28–2.67) and 1.53 (95% CI: 1.2–1.96) respectively], with marginal and no significance in the Sample 2 MPX and SPX families, respectively (Table 3).
Discussion
In this study, we provide support for a role of the DLX1/DLX2 genes in the susceptibility or cause of ASDs. Evidence for association of 5 Tag SNPs in the DLX1/DLX2 genes, namely rs788172, rs788173, rs813720, rs4519482 and rs743605, was presented. Family-based association analysis revealed that the rs4519482-T allele consistently showed significant association in the 138 Sample 1 MPX families, 169 Sample 2 MPX and the combined group of 307 MPX families, as well as in 306 SPX families. We also showed that the rs788172-G, rs788173-G and rs813720-G alleles were associated with ASD in the Sample 2 MPX families, whereas the rs743605-G allele was associated with ASD in the Sample 1 MPX families, with these four SNPs all showing significant association in the combined MPX family cohort. Interestingly, Hamilton et al22 reported an increase in the frequency of the rs743605-G allele in the sample of AGRE families they examined when compared with a control group of samples (P=0.04). We also identified a risk allele-containing haplotype, rs788172-G rs788173-G rs813720-G rs4519482-T rs743605-G, that was highly significantly over transmitted from parents to autistic children in all the families studied, with the significance in MPX families (Pcor=0.00007) being much greater than that in SPX families. We suggest that the two sub-blocks in this major haplotype block may be linked to different functional polymorphisms in the different cohorts of families.
To the best of our knowledge, this is the first comprehensive Tag SNP-based study covering the DLX1/DLX2 region using two different sets of MPX family samples. The Sample 1 MPX families were obtained from AGRE and the majority is from the United States. The Sample 2 MPX and all SPX families were recruited by ASD-CARC through an on-line Research Registry and by collaborating clinical geneticists whose clinics include a large number of individuals with definitively diagnosed ASDs. These families are predominantly from Canada, although some are also from the United States.
Many factors may contribute to a failure of replication in association studies of complex genetic disorders,35, 36 including study design, sample size, recruiting strategies, power issues, and sources of true variability among populations. Consequently, our study illustrating the association of ASD to Tag SNPs in the DLX1/DLX2 genes in two cohorts of MPX families, as well as SPX families, recruited using different strategies and different subject sources across North America, supports the DLX1/DLX2 genes as contributing factors to ASD etiology. We suggest that there are two functional polymorphisms in the DLX1/2 genes, with one present in each of the haplotype sub-blocks, and that the different family cohorts have different proportions of the functional polymorphisms. The Sample 2 families appear to have a higher proportion of a functional allele in the DLX1 gene and the Sample 1 families have a higher proportion of a functional allele in the DLX2 gene, reflecting the sub-blocks that are most strongly associated with these respective families. On the basis of this hypothesis, the SPX families would have a higher proportion of families with a functional polymorphism in the DLX2 gene.
The Homeobox-containing DLX gene complexes are of interest because they regulate the development of a subset of cortical and striatal neurons. The Dlx2 gene has been shown to be involved in forebrain development in the mouse, regulating the migration and differentiation of neurons in the subcortical telencephalon.37 DLX2 is one of the major transcription factors expressed in GABAergic neurons of the neocortex.38 Among neurotransmitter systems, some of the strongest evidence supports defects in the GABAergic inhibitory system in autism.39 Unlike pyramidal neurons, GABAergic interneurons migrate tangentially for considerable distances, and thus may be susceptible to genetic or environmental factors acting during early development.40 Thus the DLX gene complexes, which are involved in the differentiation and migration of GABAergic interneurons, can be added to the GABA receptor genes as contributing to the autism phenotype. Furthermore, the Dlx genes regulate the expression of the X-linked Arx gene in basal ganglia progenitor cell populations,41 and a subset of patients with a mutation of ARX gene have autistic features.42, 43 Both the neuropathological findings, which indicate altered organization of specific regions of the brain44, 45, 46 and the co-occurrence of autism with a number of neurological and cognitive disorders, including epilepsy,8 support a role for the DLX gene complexes in interneuron development and GABAergic-mediated pathophysiological changes that are central components of autism.47 We had only a small group of individuals (n=32) with confirmed seizures. There was increased transmission of the risk alleles and risk haplotype to affected children in this group of families, although it was much less significant than the overall group of families (data not presented).
Testing for association in SPX families revealed increased transmission of only the rs4519482-T allele (P=0.033) in single SNP analyses and over transmission of the GGGTG haplotype (P=0.012), for which the significance was lower than that in the MPX families. As most SPX families have relatively small sib-ships, it is possible that this increased transmission is restricted to those families that are ‘misclassified' as SPX secondary to reduced penetrance (may only be 10–20%) and small family size (many families had only one or two children). Furthermore, a finding of significant association or a trend towards association with autism in the SPX families strengthens the relevance of our findings, because families with other sporadic genetic causes, as well as those where environmental factors predominate, will reduce the significance substantially.
There are some important limitations to this study. First, we did not have samples from many of the unaffected siblings in the MPX cohorts. For the AGRE families, when we purchased the samples, we chose families in which there were few or no unaffected males. As a result, there could be a bias in the types of families that were included. This may account for the differences in the DLX1/2 SNPs that were found to be associated with autism in the two MPX cohorts. If the alternative alleles and haplotypes are significantly transferred to the unaffected siblings, this would strengthen our interpretation. Second, it will be important to obtain more complete information about epilepsy and seizures from each participating family so that we can determine whether there is, indeed, an association of DLX1/DLX2 haplotypes with this quantitative trait or other traits, such as head circumference or hearing loss, which can be used to stratify the families further. Finally, rare variants in the DLX1/2 genes will be important to examine as they may lead to the identification of the functional polymorphisms that are responsible for the associations with autism seen in the two MPX family cohorts. Despite these limitations, we believe that the findings do support a role for the DXL1/DLX2 genes in autism.
To conclude, we hypothesize that functional variants in the DLX1/DLX2 genes linked with the common risk haplotype found in this study, together with other vulnerability genes and epigenetic and environmental factors, contribute to the development of autism. A search for such functional variants in the DLX1/DLX2 genes is warranted.
Acknowledgments
This work was supported by a CIHR-Interdisciplinary Health Research Team grant (no. 43820, PI: JJAH) (www.AutismResearch.com), and an OMHF grant (PI: JJAH). NN and AW were trainees with the CIHR/NAAR STIHR ASPIRE (Autism Spectrum Interdisciplinary REsearch)/Training Program (PI: JJAH) (www.AutismTraining.ca). We thank all members of the ASD-CARC team for their support. We are very grateful to the families who participated in this research through our on-line Research Registry (www.AutismResearch.com). We acknowledge the resources provided by the AGRE (Autism Genetics Resource Exchange) consortium and the participating AGRE families. The AGRE is a program of Cure Autism Now and is supported, in part, by grant MH64547 from the National Institute of Mental Health to Daniel H Geschwind (PI).
References
- Bailey A, Le CA, Gottesman I, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142:74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- Bolton P, Macdonald H, Pickles A, et al. A case-control family history study of autism. J Child Psychol Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- Hughes C, Plumet MH, Leboyer M. Towards a cognitive phenotype for autism: increased prevalence of executive dysfunction and superior spatial span amongst siblings of children with autism. J Child Psychol Psychiatry. 1999;40:705–718. [PubMed] [Google Scholar]
- Abramson RK, Ravan SA, Wright HH, et al. The relationship between restrictive and repetitive behaviors in individuals with autism and obsessive compulsive symptoms in parents. Child Psychiatry Hum Dev. 2005;36:155–165. doi: 10.1007/s10578-005-2973-7. [DOI] [PubMed] [Google Scholar]
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry. 1997;154:185–190. doi: 10.1176/ajp.154.2.185. [DOI] [PubMed] [Google Scholar]
- Tuchman R, Rapin I. Epilepsy in autism. Lancet Neurol. 2002;1:352–358. doi: 10.1016/s1474-4422(02)00160-6. [DOI] [PubMed] [Google Scholar]
- Polleux F, Lauder JM. Toward a developmental neurobiology of autism. Ment Retard Dev Disabil Res Rev. 2004;10:303–317. doi: 10.1002/mrdd.20044. [DOI] [PubMed] [Google Scholar]
- Cline H. Synaptogenesis: a balancing act between excitation and inhibition. Curr Biol. 2005;15:R203–R205. doi: 10.1016/j.cub.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, et al. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, et al. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Stuhmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabionet R, Jaworski JM, Ashley-Koch AE, et al. Analysis of the autism chromosome 2 linkage region: GAD1 and other candidate genes. Neurosci Lett. 2004;372:209–214. doi: 10.1016/j.neulet.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Nabi R, Zhong H, Serajee FJ, Huq AH. No association between single nucleotide polymorphisms in DLX6 and Piccolo genes at 7q21–q22 and autism. Am J Med Genet B Neuropsychiatr Genet. 2003;119:98–101. doi: 10.1002/ajmg.b.10012. [DOI] [PubMed] [Google Scholar]
- Hamilton SP, Woo JM, Carlson EJ, Ghanem N, Ekker M, Rubenstein JL. Analysis of four DLX homeobox genes in autistic probands. BMC Genet. 2005;6:52. doi: 10.1186/1471-2156-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, et al. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69:327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, et al. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- Cohen IL. Criterion-related validity of the PDD Behavior Inventory. J Autism Dev Disord. 2003;33:47–53. doi: 10.1023/a:1022278420716. [DOI] [PubMed] [Google Scholar]
- Cohen IL, Schmidt-Lackner S, Romanczyk R, Sudhalter V. The PDD behavior inventory: a rating scale for assessing response to intervention in children with pervasive developmental disorder. J Autism Dev Disord. 2003;33:31–45. doi: 10.1023/a:1022226403878. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Ott J. Methods of analysis and resources available for genetic trait mapping. J Hered. 1999;90:68–70. doi: 10.1093/jhered/90.1.68. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19 Suppl 1:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Zhao JH. 2LD, GENECOUNTING and HAP: computer programs for linkage disequilibrium analysis. Bioinformatics. 2004;20:1325–1326. doi: 10.1093/bioinformatics/bth071. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Riley B. Linkage studies of schizophrenia. Neurotox Res. 2004;6:17–34. doi: 10.1007/BF03033293. [DOI] [PubMed] [Google Scholar]
- Ackerman H, Usen S, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP. A comparison of case-control and family-based association methods: the example of sickle-cell and malaria. Ann Hum Genet. 2005;69:559–565. doi: 10.1111/j.1529-8817.2005.00180.x. [DOI] [PubMed] [Google Scholar]
- Bartlett CW, Gharani N, Millonig JH, Brzustowicz LM. Three autism candidate genes: a synthesis of human genetic analysis with other disciplines. Int J Dev Neurosci. 2005;23:221–234. doi: 10.1016/j.ijdevneu.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de AM, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Dhossche D, Applegate H, Abraham A, et al. Elevated plasma gamma-aminobutyric acid (GABA) levels in autistic youngsters: stimulus for a GABA hypothesis of autism. Med Sci Monit. 2002;8:R1–R6. [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Cobos I, Broccoli V, Rubenstein JL. The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain. J Comp Neurol. 2005;483:292–303. doi: 10.1002/cne.20405. [DOI] [PubMed] [Google Scholar]
- Turner G, Partington M, Kerr B, Mangelsdorf M, Gecz J. Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am J Med Genet. 2002;112:405–411. doi: 10.1002/ajmg.10714. [DOI] [PubMed] [Google Scholar]
- Stromme P, Mangelsdorf ME, Scheffer IE, Gecz J. Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev. 2002;24:266–268. doi: 10.1016/s0387-7604(02)00079-7. [DOI] [PubMed] [Google Scholar]
- Steffenburg S. Neuropsychiatric assessment of children with autism: a population-based study. Dev Med Child Neurol. 1991;33:495–511. doi: 10.1111/j.1469-8749.1991.tb14915.x. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Creel D, Realmuto G, et al. Electroretinograms in autism: a pilot study of b-wave amplitudes. Am J Psychiatry. 1988;145:229–232. doi: 10.1176/ajp.145.2.229. [DOI] [PubMed] [Google Scholar]
- Gillberg C, Steffenburg S, Jakobsson G. Neurobiological findings in 20 relatively gifted children with Kanner-type autism or Asperger syndrome. Dev Med Child Neurol. 1987;29:641–649. doi: 10.1111/j.1469-8749.1987.tb08506.x. [DOI] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 2004;27:400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]