Abstract
Noise-induced hearing loss (NIHL) is one of the most important occupational health hazards. Millions of people worldwide are exposed daily to harmful levels of noise. NIHL is a complex disease resulting from an interaction between genetic and environmental factors. Although the environmental risk factors have been studied extensively, little is known about the genetic factors. Heat-shock proteins (HSPs) are induced after exposure to severe noise. When first induced by exposure to moderate sound levels, they can protect the ear from damage from excessive noise exposure. This protection is highly variable between individuals. An association of HSP70 genes with NIHL has been described by Yang et al (2006) in a Chinese sample set of noise-exposed workers. In this study, three polymorphisms (rs1043618, rs1061581 and rs2227956) in HSP70-1, HSP70-2 and HSP70-hom, respectively, were genotyped in 206 Swedish and 238 Polish DNA samples of noise-exposed subjects and analyzed. One SNP, rs2227956 in HSP70-hom, resulted in a significant association with NIHL in both sample sets. In addition, rs1043618 and rs1061581 were significant in the Swedish sample set. Analysis of the haplotypes composed of the three SNPs revealed significant associations between NIHL and haplotype GAC in both sample sets and with haplotype CGT in the Swedish sample set. In conclusion, this study replicated the association of HSP70 genes with NIHL in a second and third independent noise-exposed sample set, hereby adding to the evidence that HSP70 genes may be NIHL susceptibility genes.
Keywords: noise-induced hearing loss, HSP70, association study, replication
Introduction
Noise is one of the most widespread sources of environmental stress in living environments.1 Regular exposure to continuous noise or exposure to a single acoustic overstimulation can lead to noise-induced hearing loss (NIHL). Nowadays, millions of people worldwide are exposed daily to harmful levels of noise, rendering NIHL one of the most important occupational hazards.2 NIHL is a complex form of hearing loss; environmental and genetic factors interact and define a person's sensitivity to noise. Obviously, noise is the most important environmental factor involved in NIHL. It causes both mechanical and metabolic damage to the ear.3 In addition, chemicals such as organic solvents and individual factors such as smoking, high blood pressure and cholesterol levels influence the development of NIHL.4, 5, 6, 7, 8
Animal studies have confirmed the assumption that genetic factors influence the susceptibility to NIHL.9 Knockout mice, such as Pmca2−/−,10 Cdh23+/−,11 Sod1−/−12 and Gpx1−/−,13 are more susceptible to noise than their wild-type littermates. Mouse strains with age-related hearing impairment and spontaneously hypertensive rats are more sensitive to noise than other mouse strains and normotensive rats, respectively.14, 15, 16, 17
In humans, association studies have identified KCNE1,18 GSTM119 and CAT20 as putative NIHL susceptibility genes. However, hitherto a specific causative variant was either not yet identified (CAT)20 or has not yet been replicated in an independent population (KCNE1 and GSTM1).18, 21 Heat-shock proteins (HSPs) are a group of conserved proteins expressed ubiquitously in cells under both physiological and pathological circumstances. Under physiological conditions, HSPs function as molecular chaperones and assist in synthesis, folding, assembly and intracellular transport of many proteins. Expression of HSPs is increased under stressful conditions such as heat and other stress stimuli, including ischemia, viral infection, toxic compounds and many other.22 The HSP70 family (70 kD HSP) is probably the most predominant among all HSP proteins. HSP70s perform housekeeping and quality control functions in the cell.23 The human HSP70 family is composed of three genes: HSP70-1 (HSPA1A; OMIM:140550), HSP70-2 (HSPA1B; OMIM:603012) and HSP70-hom (HSPA1L; OMIM:140559).24 HSP70-1 and HSP70-2 genes encode an identical protein product of 641 amino acids. HSP70-1 is constitutively expressed at low levels. The expression of both HSP70-1 and HSP70-2 is heat inducible. HSP70-hom encodes a more basic protein of 641 amino acids that is highly related to HSP70-1, which is not heat inducible.24
HSPs are induced in the cochlea after acoustic overstimulation.25, 26 When first induced by moderate noise levels, HSPs have been shown to protect the cochlea from subsequent damage after severe noise exposure, although a noticeable individual variability exists.27, 28, 29 In addition, geranylgeranylacetone, an HSP inducer, protects against noise trauma in the guinea pig cochlea.30 Moreover, mice lacking heat-shock factor 1, the major transcription factor that regulates stress-inducible HSP expression, exhibit a significantly decreased recovery of a temporary threshold shift following noise overstimulation.31 Finally, a significant association was found between NIHL and two haplotypes composed of three polymorphisms located in the three HSP70 genes (rs1043618, rs1061581 and rs2227956) in a Chinese population of automobile workers.32
In this study, we investigated whether we could replicate the associations that were observed between haplotypes of the HSP70 genes and NIHL32 in a Swedish and Polish sample set.
Materials and methods
Subjects
Swedish sample set
A detailed description of the Swedish sample set can be found elsewhere.21, 33 In brief, 1261 male noise exposed workers from the mid-western part of Sweden were collected. They were divided into nine categories (three age-ranges, below 35, 35–50 and above 50 years, and three occupational noise exposure categories, ≤85 dBA, 86–91 dBA and ≥92 dBA, all leq, 8 h, 5 days a week). From each category, the 10% most resistant and the 10% most sensitive persons were selected by using the hearing threshold level (HTL) at 3 kHz of the left ear. 3 kHz was preferred for the selection of susceptible individuals over 4 or 6 kHz. Increase in damage leads to a widening of the initial 4–6 kHz notch to lower frequencies (ISO 1999 – International Organization for Standardization, 1990), and the HTL at 3 kHz continues to increase over a longer period of time.34 This was helpful because the majority of the Swedish subjects (79%) had been exposed to noise for 20–30 years or more. In addition, the ISO 1999 norm shows that individuals who have been exposed to noise (≥90 dBA) for 20 years or more have a higher HTL at 3 kHz than at 4 and 6 kHz in the 0.1 fractile. Blood samples were taken from a total of 218 subjects. Samples that had previously been removed after another genotyping effort (unpublished results), because they were indicated as genetic outliers by the programs CHECKHET (http://www.smd.qmul.ac.uk/statgen/dcurtis/software.html) and GRR,35 were also omitted from this study. A total of 206 samples, consisting of 98 noise susceptible and 108 noise-resistant subjects, were used for further analysis.
Polish sample set
Information concerning the audiometric status, noise exposure and exposure to chemicals was gathered from 3860 Polish workers from different industries, including a coal mine, an electric power station, a dockyard, a glass bottle factory and a lacquer and paint factory. An inclusion criterion for this study was an exposure to noise of at least 3 years. Subjects with a history of middle ear disease, conductive hearing loss or skull trauma and subjects with a family history of hearing loss were excluded. Unlike for the selection in the Swedish population, HTLs at 4 and 6 kHz, the two frequencies that are most easily affected by NIHL, were evaluated. In former genetic studies on these noise-exposed workers, resistant and sensitive subjects were selected using a Z-score based on the ISO 1999 (ISO 1999 – International Organization for Standardization, 1990)36 leading to a selection of 347 sensitive and 338 resistant subjects.20 Age and noise exposure level in this initial selection were not equally distributed between resistant and sensitive workers. This resulted in significant effects of the interaction between age and noise exposure level and their quadratic effects. These differences and interactions in age and noise exposure between resistant and sensitive workers were corrected for in the statistical analysis. Although this approach is valid, it led to a complex statistical model of which the results were difficult to interpret. That is why we opted to perform a stricter and matched selection of resistant and sensitive subjects for this study to facilitate the statistical analysis and the interpretation of the results. Firstly, additional exclusion criteria were applied. Only male subjects were included. Subjects with a history of extended treatment with aminoglycosides were excluded. The duration of exposure to noise had to exceed [(age * 0.666)–20 years] and finally, no subjects were included that had been exposed to noise in a previous workplace longer than 15 years. The remaining group of 3390 Polish workers was divided into nine categories according to the duration of the exposure (three levels: below 15, 15–25 and above 25 years of exposure) and the intensity of the noise exposure (three levels: ≤85 dBA, 86–91 dBA and ≥92dBA, all leq, 8 h, 5 days a week). Subsequently, the mean left ear HTL at 4 and 6 kHz was calculated for each subject. Within each of the nine categories, the 20% most resistant and the 20% most sensitive persons were selected. At this level, additional exclusion criteria were applied to the sensitive subjects only. These could not have a history of meningitis, aminoglysoside treatment or acoustic trauma. In addition, an asymmetry between the right and left ear of more than or equaling 40 dB was an exclusion factor. Finally, they should not have been exposed to noise in a previous workplace for more than 5 years. For genetic analysis, we were limited to the subjects of whom DNA was collected, being the subjects who had been selected as resistant or sensitive subjects using the previous selection procedure based on the ISO 1999. Subsequently, for each resistant subject, a matched sensitive subject for factory and age (+/−10 years) was selected. This selection procedure resulted in 119 pairs of samples, 119 resistant subjects and 119 sensitive subjects.
Genotyping
Genomic DNA was extracted from blood samples using standard procedures. The three SNPs, rs1043618 in HSP70-1 (SNP1), rs1061581 in HSP70-2 (SNP2) and rs2227956 in HSP70-hom (SNP3), were genotyped using ABI TaqMan® SNP genotyping assays (rs1043618: C_11917510_10; rs1061581: custom ABI TaqMan® SNP genotyping assay; rs2227956: C_25630755_10; Applied Biosystems, Foster City, CA, USA) on a Roche LightCycler®480 system (Roche, Basel, Switzerland). The ABI PRISM® SNaPshot™ Multiplex kit was used for validation purposes according to the manufacturer's instructions. PCR reactions were performed using standard procedures. Primer sequences, PCR annealing temperatures and Taq DNA polymerase (Invitrogen Life Technologies, San Diego, CA, USA) concentrations are listed in Table 1. The PCR products for SNaPshot™ analysis were separated on an ABI PRISM® 3130xl Genetic analyzer (Applied Biosystems) and the results were analyzed using ABI PRISM® GeneMapper™ Software Version 3.0 (Applied Biosystems).
Table 1. SNP details and PCR conditions for SNaPshot analysis.
| Gene | SNP | rs number | Nucleotide change | Primer | Primer sequence | Taq concentration (U μl−1) | PCR annealing temperature (°C) |
|---|---|---|---|---|---|---|---|
| HSP70-1 | 1 | rs1043618 | C/G | PCR forward | TCCACTACCTTTTTCGAGAG | 0.0250 | 57.3 |
| PCR reverse | GGTTCCCTGCTCTCTGTC | ||||||
| SNaPshot | CCAGCCCCCAATCTCAGAGC | ||||||
| HSP70-2 | 2 | rs1061581 | A/G | PCR forward | CATCGACTTCTACACGTCCA | 0.0250 | 57.4 |
| PCR reverse | CAAAGTCCTTGAGTCCCAAC | ||||||
| SNaPshot | GTCGCGCCCGTTGAAGAAGTC | ||||||
| HSP70-hom | 3 | rs2227956 | C/T | PCR forward | ACCAAGCAGACACAGATTTT | 0.0125 | 54.7 |
| PCR reverse | TCAAACCTTCATCACTCACA | ||||||
| SNaPshot | GTATTCTCAATGTCACAGCCA |
Statistical analysis
Hardy–Weinberg equilibrium was checked for the three SNPs using a χ2-test for goodness-of-fit (HWE version 1.2).37 Haploview was used to investigate the LD pattern between the SNPs (http://www.broad.mit.edu/mpg/haploview/). Association tests were performed using conditional stepwise backward logistic regression in the Polish matched sample set and regular stepwise backward logistic regression in the Swedish sample set. In both methods, the affection status (resistant and sensitive) was regressed on the genotypes and the interaction of the genotype with noise exposure level, correcting for age and noise exposure level. We assumed an additive genetic model where counts of a reference allele are treated as a continuous covariate. All statistical analyses were performed using SAS (SAS 9.1.3 for Windows, SAS Institute Inc., Cary, NC, USA).
For haplotype analysis, haplotypes and their likelihood were inferred by SAS Genomics using the haplotype procedure. For haplotype association testing, inferred haplotypes were weighted by their likelihood as estimated by SAS. The statistical haplotype analysis was performed using the unconditional logistic regression model described above. Each haplotype was tested against all other haplotypes.
Results
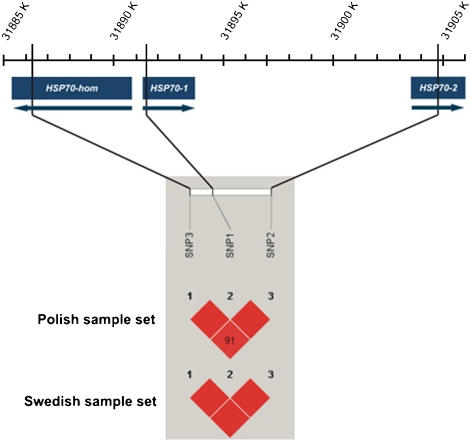
In this study, three SNPs located in three genes of the HSP70 family, rs1043618 in HSP70-1, rs1061581 in HSP70-2 and rs2227956 in HSP70-hom, were genotyped in 206 Swedish and 238 Polish noise-exposed workers. All three SNPs were in HWE and were in high LD with each other in both populations (Figure 1).
Figure 1.
Positions of HSP70-1, HSP70-2 and HSP70-hom and SNPs 1–3 on chromosome 6 (NT_007592.14). The black blocks * indicate the LD between the SNPs for the Polish and Swedish population. *Red blocks in html version.
Single SNP analysis
Table 2 contains genotype frequencies and the P-values resulting after statistical analysis of the single SNPs in the Polish and Swedish sample sets. None of the analyzed SNPs showed a significant interaction between genotype and exposure level. For the Swedish sample set, single SNP analysis of all three SNPs resulted in a significant P-value, P=0.037, P=0.033 and P=0.010, respectively. For SNP 1, genotype GG was more frequent in resistant samples (45.4%) compared to sensitive samples (31.6%), whereas genotype CC and CG were more frequent in the sensitive subjects (OR=0.61; 95% CI=0.39–0.97). For SNP 2, genotype AA appeared more often in resistant samples (40.7%) compared to sensitive samples (24.5%), whereas the sensitive subjects were more likely to be heterozygous or GG homozygous (OR=1.63; 95% CI=1.04–2.55). For SNP 3, five of the resistant samples were CC homozygotes (4.6%), whereas this genotype was not found among the sensitive subjects. Genotype TT appeared more often among the sensitive than among the resistant subjects (65.3 versus 50.0%; OR=2.09; 95% CI=1.19–3.67). In the Polish sample set, only SNP 3 showed a significant genotype effect of 0.048. For this SNP, genotype CC was found four times in the resistant group (3.4%) compared to only a single time in the sensitive group (0.8%). Again, genotype TT was more frequent in the sensitive group (79.8 versus 68.1%; OR=1.75; 95% CI=1.00–3.04).
Table 2. Single SNP analysis of three HSP70 SNPs in the Swedish and Polish sample sets.
| Sweden | Poland | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotypes | Resistant No. (%) | Sensitive No. (%) | P-value | OR (95% CI) | Resistant No. (%) | Sensitive No. (%) | P-value | OR (95% CI) |
| SNP 1 | ||||||||
| rs1043618 | ||||||||
| CC | 7 (6.5) | 11 (11.2) | 0.037 | 0.61 (0.39–0.97) | 12 (10.1) | 14 (11.8) | 0.979 | 0.99 (0.65–1.51) |
| CG | 44 (40.7) | 51 (52.0) | 58 (48.7) | 58 (48.7) | ||||
| GG | 49 (45.4) | 31 (31.6) | 46 (38.7) | 44 (37.0) | ||||
| SNP 2 | ||||||||
| rs1061581 | ||||||||
| AA | 44 (40.7) | 24 (24.5) | 0.033 | 1.63 (1.04–2.55) | 43 (36.1) | 37 (31.1) | 0.345 | 1.21 (0.82–1.80) |
| AG | 45 (41.7) | 55 (56.1) | 56 (47.1) | 61 (51.3) | ||||
| GG | 11 (10.2) | 13 (13.3) | 15 (12.6) | 18 (15.1) | ||||
| SNP 3 | ||||||||
| rs2227956 | ||||||||
| CC | 5 (4.6) | 0 (0) | 0.010 | 2.09 (1.19–3.67) | 4 (3.4) | 1 (0.8) | 0.048 | 1.75 (1.00–3.04) |
| CT | 39 (36.1) | 27 (27.5) | 32 (26.9) | 22 (18.5) | ||||
| TT | 54 (50.0) | 64 (65.3) | 81 (68.1) | 95 (79.8) | ||||
P-values that are significant at the 5% level are indicated in bold. All results are adjusted for age and noise exposure level.
Haplotype analysis
Haplotypes were inferred based on observed genotypes by SAS Genomics using the haplotype procedure. Five different haplotypes were identified (Table 3; CGT, GAC, GAT, GGC and GGT). Haplotype GGC was absent in the Swedish samples. In the latter sample set, two haplotypes, CGT and GAC, were found to be associated with NIHL (P=0.049 and P=0.017, respectively). Carriers of haplotype GAC had an almost twofold decreased odds of developing NIHL after noise exposure (OR=0.53; 95% CI=0.32–0.89) compared to all other haplotypes, whereas carriers of haplotype CGT had a 1.5 times increased odds (OR=1.53; 95% CI=1.00–2.35). In the Polish sample set, the haplotype GAC was also significantly associated with NIHL, P=0.030. Again, a twofold decreased odds was observed (OR=0.54; 95% CI=0.31–0.94). Analysis of haplotype GAT in the Polish sample set resulted in a significant interaction P-value of P=0.050, indicating that a significant difference in haplotype distribution was observed between sensitive and resistant persons for the different noise exposure levels. In other words, a differential effect of the haplotype on the noise sensitivity according to the noise exposure level may exist. In this case, odds ratios were calculated for three different noise exposure levels (81, 87 and 95 dB, which were the mean exposure levels of each noise exposure stratum). The odds ratios indicated an increased odds of haplotype GAT in the low noise exposure level group (OR=1.27; 95% CI=0.99–1.63), whereas an opposite effect, a decreased odds, was seen in the high noise exposure level group (OR=0.71; 95% CI=0.45–1.11); although, these effects did not reach significance.
Table 3. Haplotype analysis in the Swedish and Polish sample sets.
| Haplotypes | Resistant No. (%) | Sensitive No. (%) | P-value | OR (95% CI) | P-value of interaction haplotype and noise exposure level | OR (95% CI) per noise exposure level |
|---|---|---|---|---|---|---|
| Sweden | ||||||
| CGT | 29 (27.2) | 36 (37.2) | 0.049 | 1.53 (1.00–2.35) | ||
| GAC | 25 (23.0) | 14 (14.1) | 0.017 | 0.53 (0.32–0.89) | ||
| GAT | 42 (39.1) | 38 (38.9) | 0.948 | 0.99 (0.66–1.48) | ||
| GGC | 0 (0) | 0 (0) | — | — | ||
| GGT | 5 (4.2) | 5 (4.6) | 0.752 | 1.17 (0.45–3.03) | ||
| Poland | ||||||
| CGT | 42 (35.2) | 44 (37.0) | 0.825 | 1.04 (0.72–1.52) | ||
| GAC | 19 (16.3) | 11 (9.6) | 0.030 | 0.54 (0.31–0.94) | ||
| 81 dB: 1.27 (0.99–1.63) | ||||||
| GAT | 54 (45.5) | 58 (48.6) | — | — | 0.050 | 87 dB: 0.99 (0.81–1.21) |
| 95 dB: 0.71 (0.45–1.11) | ||||||
| GGC | 1 (0.6) | 1 (0.5) | 0.965 | 0.95 (0.09–10.47) | ||
| GGT | 2 (1.6) | 5 (4.2) | 0.119 | 2.58 (0.79–8.45) |
P-values that are significant at the 5% level are indicated in bold. All results are adjusted for age and noise exposure level.
Discussion
In this study, three SNPs located in three genes of the HSP70 family were analyzed in a Swedish and Polish sample set. These polymorphisms had previously been investigated in a Chinese population of noise-exposed automobile workers32 where significant associations with NIHL had been found. This renders HSP70 the second gene whose association with NIHL has been replicated in independent populations, besides CAT.20 Replication in independent sample sets is crucial to confirm susceptibility genes for complex diseases. Our results indicate that HSP70 genes may be valid NIHL susceptibility genes as significant associations were observed in three independent populations.
Yang et al32 did not detect significant associations with NIHL using single SNP analysis, but found only significant associations between two haplotypes, GGC and GGT, and NIHL. In this study, significant results were obtained after single SNP analysis. All three SNPs were significantly associated with NIHL in the Swedish sample set, whereas SNP 3 resulted in significant P-values in the Polish sample set. Also after haplotype analysis, differences were observed when compared to the Chinese study. In our study, haplotype CGT was significant in the Swedish sample set and haplotype GAC in the Swedish and Polish sample sets, whereas the haplotypes that were associated with NIHL in the Chinese population (GGC and GGT) were infrequent or absent in the Swedish and Polish population. These differences may be due to the ethnic differences between the sample sets. It is well known that the minor allele frequencies (MAF) of SNPs can differ between populations of different ancestry. For SNP 1, the MAF for the HapMap European population is comparable with the MAF for the HapMap Chinese population (0.342 and 0.311, respectively; www.hapmap.org). For SNP 3, only a small difference exists (0.292 versus 0.189). However, for SNP 2, G is the minor allele with a MAF of 0.289 in the European population, whereas A is the minor allele with a MAF of 0.477 in the Asian population, as reported on the NCBI website (http://www.ncbi.nlm.nih.gov/). Most importantly, to confirm an association of a gene with a disease, it is not necessary that identical SNPs are associated in all populations under study. Different SNPs associated in different populations, but within the same gene, can be regarded as a replication.38
Between the two European sample sets, we observed some striking similarities as well. SNP3 was significant in both sample sets with an increased odds for NIHL of the T-allele in both sample sets. In addition, haplotype GAC showed a protective effect in both sample sets, with a twofold decreased odds of developing NIHL.
Several tests were performed in this association study. As a consequence, one could argue that a multiple testing correction should be applied. However, no consensus exists on how to perform such correction, especially when the studied SNPs are in LD with each other, whereas replication of findings in several independent sample sets is nowadays believed to be more important than obtaining highly significant P-values.38 Therefore, this study based its conclusion rather on evidence of replication than on the size of the P-values.
SNP 1 of the HSP70-1 gene is positioned in the 5′ untranslated region, whereas SNP 2 is a synonymous coding SNP of HSP70-2. SNP 3 of HSP70-hom is a nonsynonymous variation; methionine is substituted for a threonine. Recently, a significant association between the C-allele of SNP 3 and increased levels of serum HSP70 has been observed.39 This interesting finding is consistent with the results of our study, as we observed a protective effect for the C-allele in both populations. Moreover, only two haplotypes, GGC and GAC, contain the C-allele of SNP 3. Haplotype GGC was not observed in the Swedish sample set and was very infrequent in the Polish samples. Haplotype GAC resulted in an almost twofold decreased odds to develop NIHL in both the Swedish and Polish sample set. Increased serum levels of HSP70 may have a protective effect on hearing under noise exposure. Increased levels of HSP70 because of the C-allele of SNP 3 may also occur in the inner ear, which may protect against hair cell damage following noise exposure. The effect of this SNP on the expression or function of HSP70 may be further investigated by generating a mouse model carrying this C-allele and testing its noise resistance. When such studies would confirm the protective effect of the C-allele, this SNP would be the first identified variant with a protective effect on the development of NIHL. However, the analyzed SNPs were in high LD with each other and with SNPs in neighboring regions in which several genes were located. We have to keep in mind that these untyped SNPs may be responsible for the association with NIHL. No functional studies were performed to exclude this possibility as this was outside the scope of this study. Such studies should be performed in the future together with additional genetic studies.
NIHL is not the first condition that was associated with the HSP70 gene family. Previously, associations have been reported with autoimmune diseases,40, 41, 42, 43 Parkinson's disease,44 abacavir hypersensitivity,45 lung cancer,46 acute high altitude illness,47 sarcoidosis48, 49 ischemic stroke50 and postoperative atrial fibrillation.39, 51 This indicates that HSP70 genes act in many stress-related diseases.
In conclusion, we analyzed three SNPs located in three genes of the HSP70 family in a Swedish and Polish sample set. Significant results of the single SNP and haplotype analyses were obtained in both sample sets. These results replicate previous findings reported in a study on Chinese noise-exposed workers32 and indicate that HSP70 genes may be valid NIHL susceptibility genes.
Acknowledgments
This study was supported by a grant from the British Royal Institute for Deaf and Hard of Hearing People (RNID) to GVC and LVL and a TOP grant from the University of Antwerp to GVC.
References
- Wallenius M. The interaction of noise stress and personal project stress on subjective health. J Environ Psych. 2004;24:167–177. [Google Scholar]
- Alberti P.Noise-induced hearing loss – a global problemin Prasher D LL (ed):Noise-induced hearing loss – a global problem London; 1998. vol 1 Protection against noise, pp7–15. [Google Scholar]
- Lim DJ. Effects of noise and ototoxic drugs at the cellular level in the cochlea: a review. Am J Otolaryngol. 1986;7:73–99. doi: 10.1016/s0196-0709(86)80037-0. [DOI] [PubMed] [Google Scholar]
- Campo P, Lataye R. Noise and solvent, alcohol and solvent: two dangerous interactions on auditory function. Noise Health. 2000;3:49–57. [PubMed] [Google Scholar]
- Toppila E, Pyykko II, Starck J, Kaksonen R, Ishizaki H. Individual risk factors in the development of noise-induced hearing loss. Noise Health. 2000;2:59–70. [PubMed] [Google Scholar]
- Fechter LD. Promotion of noise-induced hearing loss by chemical contaminants. J Toxicol Environ Health A. 2004;67:727–740. doi: 10.1080/15287390490428206. [DOI] [PubMed] [Google Scholar]
- Sliwinska-Kowalska M, Zamyslowska-Szmytke E, Szymczak W, et al. Effects of coexposure to noise and mixture of organic solvents on hearing in dockyard workers. J Occup Environ Med. 2004;46:30–38. doi: 10.1097/01.jom.0000105912.29242.5b. [DOI] [PubMed] [Google Scholar]
- Ni CH, Chen ZY, Zhou Y, et al. Associations of blood pressure and arterial compliance with occupational noise exposure in female workers of textile mill. Chin Med J (Engl) 2007;120:1309–1313. [PubMed] [Google Scholar]
- Borg E, Canlon B, Engstrom B. Noise-induced hearing loss. Literature review and experiments in rabbits. Morphological and electrophysiological features, exposure parameters and temporal factors, variability and interactions. Scand Audiol Suppl. 1995;40:1–147. [PubMed] [Google Scholar]
- Kozel PJ, Davis RR, Krieg EF, Shull GE, Erway LC. Deficiency in plasma membrane calcium ATPase isoform 2 increases susceptibility to noise-induced hearing loss in mice. Hear Res. 2002;164:231–239. doi: 10.1016/s0378-5955(01)00420-8. [DOI] [PubMed] [Google Scholar]
- Holme RH, Steel KP. Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. J Assoc Res Otolaryngol. 2004;5:66–79. doi: 10.1007/s10162-003-4021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, McFadden SL, Ding DL, et al. Targeted deletion of the cytosolic Cu/Zn-superoxide dismutase gene (Sod1) increases susceptibility to noise-induced hearing loss. Audiol Neurootol. 1999;4:237–246. doi: 10.1159/000013847. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, McFadden SL, Ding DL, Lear PM, Ho YS. Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. J Assoc Res Otolaryngol. 2000;1:243–254. doi: 10.1007/s101620010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erway LC, Shiau YW, Davis RR, Krieg EF. Genetics of age-related hearing loss in mice. III. Susceptibility of inbred and F1 hybrid strains to noise-induced hearing loss. Hear Res. 1996;93:181–187. doi: 10.1016/0378-5955(95)00226-x. [DOI] [PubMed] [Google Scholar]
- Davis RR, Newlander JK, Ling X, Cortopassi GA, Krieg EF, Erway LC. Genetic basis for susceptibility to noise-induced hearing loss in mice. Hear Res. 2001;155:82–90. doi: 10.1016/s0378-5955(01)00250-7. [DOI] [PubMed] [Google Scholar]
- Harding GW, Bohne BA, Vos JD. The effect of an age-related hearing loss gene (Ahl) on noise-induced hearing loss and cochlear damage from low-frequency noise. Hear Res. 2005;204:90–100. doi: 10.1016/j.heares.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Borg E. Noise-induced hearing loss in normotensive and spontaneously hypertensive rats. Hear Res. 1982;8:117–130. doi: 10.1016/0378-5955(82)90070-3. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Carlsson PI, Ottschytsch N, et al. The contribution of genes involved in potassium recycling in the inner ear to noise-induced hearing loss. Hum Mutat. 2006;27:786–795. doi: 10.1002/humu.20360. [DOI] [PubMed] [Google Scholar]
- Rabinowitz PM, Pierce Wise J, Sr, Hur Mobo B, Antonucci PG, Powell C, Slade M. Antioxidant status and hearing function in noise-exposed workers. Hear Res. 2002;173:164–171. doi: 10.1016/s0378-5955(02)00350-7. [DOI] [PubMed] [Google Scholar]
- Konings A, Van Laer L, Pawelczyk M, et al. Association between variations in CAT and noise-induced hearing loss in two independent noise-exposed populations. Hum Mol Genet. 2007;16:1872–1883. doi: 10.1093/hmg/ddm135. [DOI] [PubMed] [Google Scholar]
- Carlsson PI, Van Laer L, Borg E, et al. The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear Res. 2005;202:87–96. doi: 10.1016/j.heares.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32:242–251. doi: 10.1007/BF00187095. [DOI] [PubMed] [Google Scholar]
- Lim HH, Jenkins OH, Myers MW, Miller JM, Altschuler RA. Detection of HSP 72 synthesis after acoustic overstimulation in rat cochlea. Hear Res. 1993;69:146–150. doi: 10.1016/0378-5955(93)90102-7. [DOI] [PubMed] [Google Scholar]
- Samson J, Sheeladevi R, Ravindran R, Senthilvelan M. Stress response in rat brain after different durations of noise exposure. Neurosci Res. 2007;57:143–147. doi: 10.1016/j.neures.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Kristiansen A, Liberman MC. Heat stress and protection from permanent acoustic injury in mice. J Neurosci. 1999;19:10116–10124. doi: 10.1523/JNEUROSCI.19-22-10116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Wang XJ, Song JS, Chen HX, Man YH. Influence of evoked HSP70 expression on hearing function of the cochlea in guinea pigs. Di Yi Jun Yi Da Xue Xue Bao. 2002;22:922–924. [PubMed] [Google Scholar]
- Altschuler RA, Fairfield D, Cho Y, et al. Stress pathways in the rat cochlea and potential for protection from acquired deafness. Audiol Neurootol. 2002;7:152–156. doi: 10.1159/000058301. [DOI] [PubMed] [Google Scholar]
- Mikuriya T, Sugahara K, Takemoto T, et al. Geranylgeranylacetone, a heat shock protein inducer, prevents acoustic injury in the guinea pig. Brain Res. 2005;1065:107–114. doi: 10.1016/j.brainres.2005.10.045. [DOI] [PubMed] [Google Scholar]
- Fairfield DA, Lomax MI, Dootz GA, et al. Heat shock factor 1-deficient mice exhibit decreased recovery of hearing following noise overstimulation. J Neurosci Res. 2005;81:589–596. doi: 10.1002/jnr.20417. [DOI] [PubMed] [Google Scholar]
- Yang M, Tan H, Yang Q, et al. Association of hsp70 polymorphisms with risk of noise-induced hearing loss in Chinese automobile workers. Cell Stress Chaperones. 2006;11:233–239. doi: 10.1379/CSC-192R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson PI, Borg E, Grip L, Dahl N, Bondeson ML. Variability in noise susceptibility in a Swedish population: the role of 35delG mutation in the Connexin 26 (GJB2) gene. Audiol Med. 2004;2:123–130. [Google Scholar]
- Taylor W, Pearson J, Mair A, Burns W. Study of noise and hearing in jute weaving. J Acoust Soc Am. 1965;38:113–120. doi: 10.1121/1.1909580. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Sliwinska-Kowalska M, Dudarewicz A, Kotylo P, Zamyslowska-Szmytke E, Pawlaczyk-Luszczynska M, Gajda-Szadkowska A. Individual susceptibility to noise-induced hearing loss: choosing an optimal method of retrospective classification of workers into noise-susceptible and noise-resistant groups. Int J Occup Med Env Heal. 2006;19:235–245. doi: 10.2478/v10001-006-0029-2. [DOI] [PubMed] [Google Scholar]
- Ott J. John Hopkins University Press: Baltimore; 1999. Analysis of human genetic linkage. [Google Scholar]
- Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal AR, Mandal K, Nyamweya S, et al. Association of Met439Thr substitution in heat shock protein 70 gene with postoperative atrial fibrillation and serum HSP70 protein levels. Cardiology. 2007;110:45–52. doi: 10.1159/000109406. [DOI] [PubMed] [Google Scholar]
- Pugliese A, Awdeh ZL, Galluzzo A, Yunis EJ, Alper CA, Eisenbarth GS. No independent association between HSP70 gene polymorphism and IDDM. Diabetes. 1992;41:788–791. doi: 10.2337/diab.41.7.788. [DOI] [PubMed] [Google Scholar]
- Favatier F, Bornman L, Hightower LE, Gunther E, Polla BS. Variation in hsp gene expression and Hsp polymorphism: do they contribute to differential disease susceptibility and stress tolerance. Cell Stress Chaperones. 1997;2:141–155. doi: 10.1379/1466-1268(1997)002<0141:vihgea>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile A, Nieto A, Mataran L, Martin J. HSP70 gene polymorphisms in ankylosing spondylitis. Tissue Antigens. 1998;51:382–385. doi: 10.1111/j.1399-0039.1998.tb02977.x. [DOI] [PubMed] [Google Scholar]
- Vargas-Alarcon G, Londono JD, Hernandez-Pacheco G, et al. Heat shock protein 70 gene polymorphisms in Mexican patients with spondyloarthropathies. Ann Rheum Dis. 2002;61:48–51. doi: 10.1136/ard.61.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Wang CK, Chen CM, et al. Analysis of heat-shock protein 70 gene polymorphisms and the risk of Parkinson's disease. Hum Genet. 2004;114:236–241. doi: 10.1007/s00439-003-1050-1. [DOI] [PubMed] [Google Scholar]
- Martin AM, Nolan D, Gaudieri S, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci USA. 2004;101:4180–4185. doi: 10.1073/pnas.0307067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusin M, Zientek H, Krzesniak M, et al. Intronic polymorphism (1541-1542delGT) of the constitutive heat shock protein 70 gene has functional significance and shows evidence of association with lung cancer risk. Mol Carcinog. 2004;39:155–163. doi: 10.1002/mc.20009. [DOI] [PubMed] [Google Scholar]
- Zhou F, Wang F, Li F, et al. Association of hsp70-2 and hsp-hom gene polymorphisms with risk of acute high-altitude illness in a Chinese population. Cell Stress Chaperones. 2005;10:349–356. doi: 10.1379/CSC-156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunia-Kubik K, Koscinska K, Suchnicki K, Lange A. HSP70-hom gene single nucleotide (+2763 G/A and +2437 C/T) polymorphisms in sarcoidosis. Int J Immunogenet. 2006;33:135–140. doi: 10.1111/j.1744-313X.2006.00584.x. [DOI] [PubMed] [Google Scholar]
- Spagnolo P, Sato H, Marshall SE, et al. Association between heat shock protein 70/Hom genetic polymorphisms and uveitis in patients with sarcoidosis. Invest Ophthalmol Vis Sci. 2007;48:3019–3025. doi: 10.1167/iovs.06-1485. [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng J, Peng J, Han S, Yu L, Nie S. Effects of polymorphisms of heat shock protein 70 gene on ischemic stroke, and interaction with smoking in China. Clin Chim Acta. 2007;384:64–68. doi: 10.1016/j.cca.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Mandal K, Torsney E, Poloniecki J, Camm AJ, Xu Q, Jahangiri M. Association of high intracellular, but not serum, heat shock protein 70 with postoperative atrial fibrillation. Ann Thorac Surg. 2005;79:865–871. doi: 10.1016/j.athoracsur.2004.08.018. [DOI] [PubMed] [Google Scholar]