Abstract
Lactase persistence is an autosomal-dominant trait that is common in European-derived populations. A basic tendency for lactase persistence to increase from the southeast to the northwest across European populations has been noted, but such trends within countries have not been extensively studied. We genotyped the C/T−13910 variant (rs4988235) that constitutes the putatively causal allele for lactase persistence (T allele representing persistence) in a general population sample of 3344 women aged 60–79 years from 23 towns across Britain. We found an overall frequency of 0.253 for the C (lactase non-persistence) allele, but with considerable gradients of decreasing frequency from the south to the north and from the east to the west of Britain for this allele. Daily sunlight was positively related to C (non-persistence) allele prevalence. However, sunlight exposure and latitude are strongly correlated, and it was not possible to identify which is the primary factor statistically underlying the distribution of lactase persistence. The C/T−13910 variant (rs4988235) was not related to drinking milk or bone health (although drinking milk itself was protective of bone health), and was essentially unrelated to a wide range of other lifestyle, health and demographic characteristics. One exception was general health being rated as being poor or fair, for which there was an odds ratio of 1.38 (1.04, 1.84) for women homozygous for the C allele; on adjustment for latitude and longitude of place of birth, this attenuated to 1.19 (0.87, 1.64). The lactase persistence variant could contribute to the examination of data for the existence of, and then statistical control for, population substructure in genetic association studies.
Keywords: lactase persistence, British Women's Heart and Health Study, bone health, population stratification, milk
Introduction
Lactose is the major carbohydrate in milk and to be absorbed it needs to be broken down by hydrolysis in the intestinal tract. Lactase, an enzyme located in the brush border of the small intestine, catalyses the hydrolysis of lactose to monosaccharides, which are absorbed and serve as a source of energy.1 Milk is the major source of energy in infancy, and lactase activity at this age is universal. Lactase activity declines after the age of weaning in most mammals, although in humans this pattern is variable. Although most populations of the world have a low prevalence of lactase persistence, Northern European populations tend to have high prevalences.2 Lactose tolerance is traditionally thought of as an autosomal-dominant genetic trait, although the actual levels of lactase in the intestinal mucosa show a trimodal distribution, with very low levels in the people homozygous for the lactase non-persistence variant.3 The selective advantage of lactase persistence in milk-producing dairy farming populations, and the consequent expansion of milk production in successful populations, could have allowed for rapid selective pressures or an inceptive niche construction event,4 leading to increases in the prevalence of a genetic variant associated with lactase persistence.5 This is supported by molecular genetic evidence, which suggests strong recent selection occurring in the vicinity of the lactase gene.6, 7
Recently, a genetic variant associated with lactase persistence was identified,8 and the use of genotype data to avoid time-consuming and potentially misleading lactose tolerance tests has rendered study of lactase persistence more straightforward.9 Previous work has presented evidence of a complete correlation between lactase persistence and a common variant located 14 kb upstream of the lactase coding region (LCT) in the MCM6 gene.8 Further to this, extensive linkage disequilibrium for 1 Mb across LCT has been reported in a North European population.10 This work places the correlated allele on an extended common haplotype background and there is redundancy with respect to the genotyping of further variants associated with lactase persistence. Therefore, genotyping of the putatively causal C/T−13910 variant (rs4988235) will effectively capture the genetic variation highlighted by previous association studies.8, 10 The T allele of this single nucleotide polymorphism (SNP) is associated with lactase persistence and its prevalence has been shown to vary across Europe, being 70–80% in Northern European populations and 5–10% in Southern European populations.6
The geographic variation in the prevalence of lactase persistence raises the possibility that C/T−13910 may also vary geographically within a country such as Britain, as part of the north–south gradient seen across much of Europe.2
Such phenotypic correlations are important for the understanding of both why such geographical patterning may exist and also how such variation in genotype frequencies can influence the interpretation of gene–disease associations, given the potential for population stratification to generate spurious allelic association.11, 12, 13, 14, 15
Methods
We have used data from the British Women's Heart and Health Study. Full details of the selection of participants and measurements have been reported earlier.16, 17 Women aged 60–79 years were randomly selected from primary care lists in 23 British towns. A total of 4286 women participated and baseline data were collected between April 1999 and March 2001.
Women were asked whether they drank milk and what type of milk they drank (never drink milk or usually drink: full cream, semi-skimmed and skimmed). They were asked whether they had ever been diagnosed by a doctor as having osteoporosis and whether they had ever fractured their hip or wrist. Details for methods for other phenotype assessment, determination of LCT C/T13910 variant (rs4988235) genotypes and the study ethics are provided in the Supplementary web methods.
Statistical analyses
Logistic regression was used to assess the association between a self-report of never drinking milk versus drinking milk with osteoporosis and fractures. Prevalences (for dichotomous variables) and means (for continuous variables) are presented by genotype. Linear and logistic regressions were used for testing differences between genotypes for continuous and dichotomous variables, respectively. Further details of the statistical models are provided in the Supplementary web methods.
Results
Of the 4278 participants who gave consent for genetic testing, 15 (5 Afro-Caribbean, 8 South Asian and 2 other) were defined by the examining nurse as not being white and were excluded from further analysis. Of the remaining 4263 women, 3553 (83%) had DNA available for genotyping, and for 3344 (94%) of these women, the genotypic primary florescence data fell into one of three distinct clusters with positive signal for at least one allele. There was no difference in mean age (68.9 (5.5) versus 69.0 (5.7) years, P=0.4)) and no difference in the prevalence of never drinking milk (2.8 versus 2.5%, P=0.6) between those with and without genotypic data. Similarly, mean longitude and latitude of place of birth measures for both place of birth and place of residence were the same in those with and without genotypic data (all P>0.7). Supplementary web table 1 (see journal website) shows the characteristics of the participants included in the analyses presented in this paper. Table 1 shows genotype and allele frequencies for LCT C/T−13910 variant (rs4988235) in this study sample. Observed frequencies matched those reported earlier for North European origin populations2 and maintained Hardy–Weinberg equilibrium in the total sample (P=0.2) and among those aged 60–69 years (P=0.2) and those aged 70–79 years (P=0.6).
Table 1. Genotype and allele frequency for rs4988235 among study participants.
| Genotype | N (proportion) | Allele | N (proportion) |
|---|---|---|---|
| TT | 1881 (0.562) | T | 4996 (0.747) |
| TC | 1236 (0.370) | Ca | 1690 (0.253) |
| CC | 227 (0.068) |
N=3344.
HWE exact test P=0.22.
The lactase non-persistence allele.
Milk consumption, bone health, lifestyle, anthropometric, vascular, metabolic and socioeconomic characteristic associations with genotype
Among women with genotypic data, 3143 (94%) responded to the question concerning milk consumption, and of these, 89 (2.8%) reported never drinking milk. Never drinking milk was not associated with socioeconomic position in childhood or adulthood, vegetarianism, frequency of consumption of fruit and vegetables, red or processed meat, type of cooking fat, physical activity or smoking (all P-values >0.5). However, women who reported never drinking milk were more likely to have osteoporosis (age-adjusted odds ratio (95% confidence interval) as compared with milk drinkers (2.07 (1.10, 3.88), P=0.01), and were more likely to have a history of having had either a hip or wrist fracture (1.74 (1.09, 2.77), P=0.02).
Table 2 shows the characteristics of participants by genotype status and the P-values for the three genetic models of association that we tested. There was no evidence that those with either one or two copies of the lactase non-persistence (C) allele were different from homozygotes for the persistence (T) allele with respect to milk consumption, osteoporosis, fractures or calcium supplementation. Anthropometric, vascular, metabolic traits, socioeconomic position, lifestyle and fertility characteristics were largely unrelated to genotype. However, women who carried either one or two lactase non-persistence (C) alleles (ie, were CT or CC) had higher HDLc than those who were homozygous for the T allele (ie, TT): mean difference of 0.04 mmol/l (95% CI: 0.01, 0.07). Women who were homozygous for the C allele (CC) reported a higher prevalence of their general health being poor or fair than all other women (CT or TT), with an odds ratio of 1.38 (1.04, 1.84). There was no difference in the effect of genotype on HDLc levels or general health when stratified by drinking milk or not (P-value for interaction=0.48 for HDLc outcome and 0.85 for general health outcome). As anticipated genotype was not associated with age, and therefore despite many of the characteristics presented in Table 2 being related to age, the lack of any association with genotype would mean that any association of genotype with these characteristics could not be explained by age effects. This is demonstrated in Supplementary Table 2 (see journal website), which presents the associations of genotype with participant characteristics after adjustment for age; the results are essentially identical to those presented in Table 2 in this paper.
Table 2. Prevalence and means (95% CI) of participant characteristics by genotype status among British women described as white by examining nurse and aged 60–79 years.
| Percent or mean (95% CI) by rs4988235 genotype | ||||
|---|---|---|---|---|
| TT | TC | CC | ||
| Number of subjects | 1881 | 1236 | 227 | P-valuea |
| Age (years) | 68.8 (68.5, 69.0) | 69.0 (68.6, 69.3) | 69.0 (68.3, 69.7) | 0.54 |
| 0.27 | ||||
| 0.53 | ||||
| Milk consumption, osteoporosis, fractures and cataracts | ||||
| Non-milk drinker (%) | 2.7 (2.0, 3.5) | 3.0 (2.2, 4.2) | 3.3 (1.6, 6.7) | 0.79 |
| 0.49 | ||||
| 0.70 | ||||
| Full-fat milk drinker (%) | 16.7 (15.0, 18.6) | 16.2 (14.2, 18.5) | 11.1 (7.5, 16.1) | 0.09 |
| 0.10 | ||||
| 0.04 | ||||
| Hip or wrist fractures (%) | 12.8 (11.4, 14.4) | 15.2 (13.3, 17.3) | 15.4 (11.3, 20.7) | 0.13 |
| 0.06 | ||||
| 0.49 | ||||
| Osteoporosis (%) | 7.6 (6.5, 8.9) | 8.4 (7.0, 10.1) | 9.7 (6.5, 14.3) | 0.47 |
| 0.22 | ||||
| 0.34 | ||||
| Using calcium supplement (%) | 1.0 (0.6, 1.5) | 0.8 (0.4, 1.5) | 0 | 0.57 |
| 0.17 | ||||
| NA | ||||
| Cataracts (%) | 12.5 (11.1, 14.1) | 13.3 (11.5, 15.3) | 15.0 (10.9, 20.2) | 0.53 |
| 0.28 | ||||
| 0.35 | ||||
| General health, other dietary factors and behaviours | ||||
| Reports general health as fair or poor (rather than good or excellent) | 28.1 (26.1, 30.2) | 25.5 (23.1, 28.0) | 33.9 (28.1, 40.3) | 0.02 |
| 0.83 | ||||
| 0.03 | ||||
| EuroQul anxiety/depression (%) | 23.2 (21.3) | 22.4 (20.2, 24.8) | 20.7 (15.9, 26.5) | 0.66 |
| 0.38 | ||||
| 0.45 | ||||
| Eats less than one portion per day fruit/vegetables (%) | 30.0 (27.6, 31.8) | 29.4 (27.0, 32.1) | 26.4 (21.1, 32.5) | 0.59 |
| 0.45 | ||||
| 0.32 | ||||
| Eats white bread as main bread (%) | 15.8 (14.2, 17.5) | 16.4 (14.4, 18.6) | 18.5 (13.9, 24.1) | 0.56 |
| 0.32 | ||||
| 0.33 | ||||
| Eats cheese at least once a day (%) | 9.6 (8.3, 11.1) | 10.0 (8.4, 11.9) | 10.7 (7.2, 15.7) | 0.85 |
| 0.58 | ||||
| 0.64 | ||||
| Eats red/processed meat more than once a day (%) | 4.3 (3.4, 5.3) | 4.8 (3.8, 6.2) | 1.8 (0.7, 4.6) | 0.07 |
| 0.51 | ||||
| 0.06 | ||||
| Uses lard as main cooking fat (%) | 5.9 (4.9, 7.1) | 5.6 (4.4, 7.0) | 3.5 (1.8, 6.9) | 0.30 |
| 0.23 | ||||
| 0.16 | ||||
| Physically inactive (%) | 19.6 (17.9, 21.5) | 19.6 (17.5, 22.0) | 18.9 (14.4, 24.6) | 0.97 |
| 0.88 | ||||
| 0.80 | ||||
| Current smoker (%) | 10.8 (9.5, 12.3) | 11.6 (9.9, 13.5) | 11.9 (8.3, 16.8) | 0.74 |
| 0.45 | ||||
| 0.72 | ||||
| Ever smoked (%) | 44.7 (42.5, 47.0) | 42.3 (39.6, 45.1) | 46.7 (40.3, 53.2) | 0.29 |
| 0.66 | ||||
| 0.39 | ||||
| Heavy alcohol consumption: ≥3 units per day (%) | 2.6 (2.0, 3.5) | 2.3 (1.6, 3.3) | 1.9 (0.7, 5.0) | 0.64 |
| 0.44 | ||||
| 0.60 | ||||
| Anthropometric measurements | ||||
| Height (mm) | 1588.0 (1585.2, 1590.7) | 1586.7 (1583.2, 1590.2) | 1582.0 (1573.5, 1590.4) | 0.41 |
| 0.23 | ||||
| 0.22 | ||||
| Leg length (mm) | 756.9 (755.0, 758.6) | 757.7 (755.4, 760.0) | 753.7 (748.0, 759.3) | 0.41 |
| 0.73 | ||||
| 0.23 | ||||
| Weight (kg) | 69.8 (69.2, 70.4) | 69.0 (68.3, 69.8) | 68.9 (67.3, 70.6) | 0.19 |
| 0.07 | ||||
| 0.31 | ||||
| BMI (kg/m2) | 27.7 (27.5, 27.9) | 27.4 (27.1, 27.7) | 27.7 (27.0, 28.4) | 0.32 |
| 0.22 | ||||
| 0.92 | ||||
| Waist-to-hip ratio | 0.819 (0.816, 0.822) | 0.819 (0.815, 0.822) | 0.820 (0.812, 0.829) | 0.96 |
| 0.88 | ||||
| 0.78 | ||||
| Vascular and metabolic measurements | ||||
| Systolic BP (mm Hg) | 148.0 (146.9, 149.2) | 146.3 (144.9, 147.7) | 146.1 (142.8, 149.3) | 0.10 |
| 0.05 | ||||
| 0.46 | ||||
| Diastolic BP (mm Hg) | 79.7 (79.2, 80.3) | 79.2 (78.6, 79.9) | 79.4 (78.0, 80.9) | 0.49 |
| 0.33 | ||||
| 0.92 | ||||
| Fasting insulin (logged mU/l) | 1.92 (1.89, 1.96) | 1.89 (1.80, 1.98) | 1.96 (1.91, 2.02) | 0.15 |
| 0.05 | ||||
| 0.15 | ||||
| Fasting glucose (logged mmol/l) | 1.78 (1.77, 1.79) | 1.79 (1.78, 1.80) | 1.79 (1.77, 1.82) | 0.39 |
| 0.17 | ||||
| 0.45 | ||||
| Fasting total cholesterol (mmol/l) | 6.63 (6.57, 6.68) | 6.66 (6.60, 6.73) | 6.60 (6.45, 6.75) | 0.63 |
| 0.69 | ||||
| 0.69 | ||||
| Fasting LDLc (mmol/l) | 4.14 (4.08, 4.19) | 4.17 (4.10, 4.23) | 4.09 (3.95, 4.22) | 0.54 |
| 0.98 | ||||
| 0.39 | ||||
| Fasting HDLc (mmol/l) | 1.64 (1.62, 1.66) | 1.68 (1.65, 1.70) | 1.69 (1.63, 1.75) | 0.01 |
| 0.004 | ||||
| 0.15 | ||||
| Fasting triglycerides (logged mmol/l) | 0.51 (0.49, 0.54) | 0.50 (0.48, 0.53) | 0.51 (0.45, 0.57) | 0.74 |
| 0.52 | ||||
| 0.93 | ||||
| Fasting CRP (logged mg/l) | 0.63 (0.58, 0.68) | 0.62 (0.56, 0.68) | 0.71 (0.56, 0.86) | 0.61 |
| 0.63 | ||||
| 0.33 | ||||
| Indicators of socioeconomic position and fertility | ||||
| Childhood manual social class (%) | 80.8 (78.9, 82.5) | 78.6 (76.2, 80.8) | 77.1 (71.2, 82.1) | 0.19 |
| 0.07 | ||||
| 0.31 | ||||
| No bathroom as a child (%) | 38.3 (36.1, 40.6) | 39.1 (36.4, 41.9) | 37.4 (31.2, 44.1) | 0.85 |
| 0.91 | ||||
| 0.72 | ||||
| No hot water as child (%) | 34.4 (32.3, 36.7) | 35.4 (32.7, 38.3) | 31.5 (25.6, 38.0) | 0.52 |
| 0.81 | ||||
| 0.32 | ||||
| Shared bedroom (%) | 56.7 (54.5, 58.9) | 55.9 (53.1, 58.7) | 53.3 (46.8, 59.7) | 0.60 |
| 0.35 | ||||
| 0.36 | ||||
| No car access as child (%) | 84.7 (83.0, 86.3) | 82.8 (80.6, 84.8) | 78.9 (73.1, 83.7) | 0.05 |
| 0.02 | ||||
| 0.05 | ||||
| Left school <15 years of age (%) | 34.2 (32.1, 36.4) | 32.8 (30.3, 35.5) | 28.2 (22.7, 34.4) | 0.17 |
| 0.09 | ||||
| 0.09 | ||||
| Adult manual social class (%) | 58.1 (55.8, 60.3) | 55.1 (52.3, 57.9) | 50.2 (43.7, 56.7) | 0.04 |
| 0.01 | ||||
| 0.05 | ||||
| No car access as adult (%) | 28.2 (26.2, 30.4) | 28.8 (26.3, 31.5) | 23.0 (17.9, 29.1) | 0.20 |
| 0.39 | ||||
| 0.09 | ||||
| Council housing (%) | 13.4 (11.9, 15.1) | 13.9 (12.0, 16.0) | 11.5 (7.9, 16.5) | 0.64 |
| 0.78 | ||||
| 0.39 | ||||
| State pension only (%) | 32.2 (30.1, 34.4) | 32.0 (29.4, 34.7) | 27.4 (21.9, 33.8) | 0.35 |
| 0.30 | ||||
| 0.15 | ||||
| Three or more siblings (%) | 38.2 (36.0, 40.5) | 36.6 (33.9, 39.4) | 36.9 (30.6, 43.6) | 0.66 |
| 0.42 | ||||
| 0.84 | ||||
| Nulliparous (%) | 9.8 (8.5, 11.3) | 9.5 (7.9, 11.4) | 7.7 (4.7, 12.1) | 0.59 |
| 0.41 | ||||
| 0.34 | ||||
| Median (IQR) number of pregnancies | 2 (2, 3) | 2 (2, 3) | 2 (2, 3) | 0.90 |
| 0.98 | ||||
| 0.72 | ||||
| Indicators of latitude, longitude and hours of sunlight | ||||
| Northing for area of birth (km north of the National grid originb) | 407.1 (398.9, 415.3) | 372.5 (362.6, 382.4) | 348.5 (326.2, 370.8) | <0.0001 |
| <0.0001 | ||||
| <0.0001 | ||||
| Easting for area of birth (km east of the National grid originb) | 394.9 (390.3, 399.5) | 414.4 (408.5, 420.3) | 436.1 (421.0, 451.2) | <0.0001 |
| <0.0001 | ||||
| 0.001 | ||||
| Mean hours of sunlight as % of total hours daylight for area of birth | 29.15 (29.06, 29.26) | 29.59 (29.45, 29.71) | 29.96 (29.62, 30.29) | <0.0001 |
| <0.0001 | ||||
| <0.0001 | ||||
P-value not in bold or italics (top value)=global heterogeneity effects using ANOVA (2 d.f.); P-value in bold (middle value)=per allele trend test (1 d.f.); P-value in italics=classic recessive model (non-persistence homozygotes versus all other participants, 1 d.f.). Supplementary Table 1 on the journal website in addition shows all three P-values for these three different genetic models adjusted for latitude (northing) and longitude (easting) of the area of birth for all phenotypes except these measures of the area of birth (this adjustment did not markedly alter any of the P-values).
National grid origin is near to Scilly Isles, which is one of the furthest southwesterly points of Britain. Higher values for northing represent more northerly areas and higher values for easting more easterly areas.
Gene, place of birth and area of residence association
The prevalence of homozygotes for the non-persistence (C) allele in the whole population was 6.8% (95% CI: 6.0, 7.7), which is similar to the prevalence among those women (N=3224) who were born in Britain, that is 6.1% (95% CI: 5.3, 7.0). The (C) allele frequency was also similar in the whole sample (0.253 (95% CI: 0.242, 0.263) and those born in Britain (0.246 (95% CI: 0.236, 0.257).
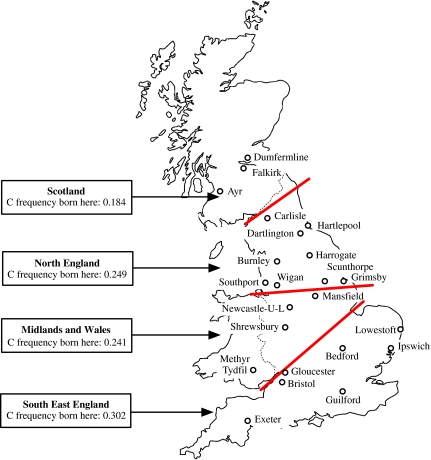
Of those women with genotype data, 3184 (95%) were born in Britain and had data on place of birth and all of these had data on area of residence. All results in this section are based on these 3184 women. Women with either one or two copies of the non-persistence (C) allele were more likely to have been born in the south of England and least likely to have been born in Scotland (Figure 1 and Table 3). A similar marked geographical variation for adult area of residence was also seen (Tables 4 and 5). The odds ratio (95% CI) of carrying a non-persistence (C) allele comparing those born in the south of England with all other areas was 1.63 (1.40, 1.91) and for comparing those living in the south of England with those living in all other areas in adulthood was 1.38 (1.19, 1.61). When both area of birth and area of adult residence were included simultaneously in a logistic regression model, the association between area of birth and possession of a minor allele remained largely unchanged (1.65 (1.33, 2.04)), whereas that with area of residence attenuated to the null (0.98 (0.80, 1.22)).
Figure 1.
C (lactase non-persistence) allele frequency of rs4988235 by area of birth for participants in British Women's Heart and Health Study.
Table 3. rs4988235 genotype and allele frequency by area of birth among women aged 60–79 years described by examining nurse as being white and who have complete data for genotype, area of birth and area of residence data.
| No (proportion) of genotype/allele by area birth | ||||
|---|---|---|---|---|
| South East England N=947 | Midlands and Wales N=684 | North England N=1168 | Scotland N=516 | |
| TT | 463 (0.490) | 394 (0.576) | 667 (0.571) | 344 (0.667) |
| TC | 395 (0.417) | 250 (0.366) | 421 (0.360) | 157 (0.304) |
| CC | 89 (0.094) | 40 (0.059) | 80 (0.068) | 15 (0.029) |
| T | 1321 (0.698) | 1038 (0.759) | 1755 (0.751) | 845 (0.819) |
| Ca | 573 (0.302) | 330 (0.241) | 581 (0.249) | 187 (0.184) |
| HW test | P=0.7 | P=0.99 | P=0.2 | P=0.7 |
| Comparisons allele frequency across areas | Likelihood ratio test for trend in allele frequency χ(1)2=37.53, P<0.0001 Likelihood ratio test for difference in allele frequency χ(3)2=44.33, P<0.0001 | |||
The lactase non-persistence allele.
N=3315.
Table 4. rs4988235 genotype and allele frequency by area of residence among women aged 60–79 years described by examining nurse as being white and who have complete data for genotype, area of birth and area of residence data.
| No. (proportion) of genotype/allele by area residence | ||||
|---|---|---|---|---|
| South East England N=1013 | Midlands and Wales N=499 | North England N=1361 | Scotland N=442 | |
| TT | 512 (0.505) | 294 (0.589) | 773 (0.568) | 289 (0.654) |
| TC | 409 (0.404) | 176 (0.353) | 503 (0.370) | 135 (0.305) |
| CC | 92 (0.091) | 29 (0.058) | 85 (0.062) | 18 (0.041) |
| T | 1433 (0.707) | 764 (0.766) | 2049 (0.753) | 713 (0.807) |
| Ca | 593 (0.293) | 234 (0.234) | 673 (0.247) | 171 (0.193) |
| HW test | P=0.4 | P=0.7 | P=0.8 | P=0.6 |
| Comparisons allele frequency across areas | Likelihood ratio test for trend in allele frequency χ(1)2=23.2, P<0.0001 Likelihood ratio test for difference in allele frequency χ(3)2=30.2, P<0.0001 | |||
The lactase non-persistence allele.
N=3315.
Table 5. Non-persistence C allele frequency by town of residence in adulthood.
| Town | No. of participants in town | No. (proportion) of minor allele frequency |
|---|---|---|
| Guildford | 201 | 118 (0.294) |
| Exeter | 105 | 183 (0.287) |
| Ipswich | 133 | 133 (0.331) |
| Lowestoft | 139 | 84 (0.302) |
| Bedford | 59 | 33 (0.280) |
| Bristol | 118 | 61 (0.259) |
| Gloucester | 136 | 75 (0.276) |
| Merthyr Tydfil | 111 | 51 (0.230) |
| Shrewsbury | 196 | 93 (0.237) |
| Mansfield | 140 | 72 (0.257) |
| Newcastle-Under-Lyme | 57 | 23 (0.202) |
| Grimsby | 162 | 85 (0.262) |
| Scunthorpe | 167 | 90 (0.270) |
| Wigan | 179 | 89 (0.249) |
| Southport | 156 | 74 (0.237) |
| Burnley | 132 | 64 (0.242) |
| Harrogate | 70 | 34 (0.243) |
| Darlington | 183 | 96 (0.262) |
| Hartlepool | 127 | 65 (0.256) |
| Carlisle | 196 | 85 (0.217) |
| Ayr | 162 | 69 (0.213) |
| Falkirk | 182 | 67 (0.184) |
| Dunfermline | 100 | 36 (0.180) |
| Comparisons allele frequency across towns of residence | Likelihood ratio test for trend (order as in first column) in allele frequency χ(1)2=31.0, P<0.0001 | |
Towns listed in the order of approximate geographical position from most southeast to most northwest.
When we examined frequency of the non-persistence (C) allele by town of residence in adulthood, we found a gradient of increasing frequency in the more southerly and easterly regions (Table 5). Because the women were born in a large number of different towns and for many towns there were only one or two women from the study born there, we are unable to present allele frequencies by town of birth. However, when we examined these associations using indicators of latitude and longitude for area of birth, there was a linear trend of women with a non-persistence (C) allele being from more southerly and easterly areas (Table 2). Women with this allele were also more likely to be from areas that experienced a greater number of hours of sunlight. The mean difference in distance north (latitude) among women with either one or two non-persistence (C) alleles as compared with homozygotes for the persistent T allele was −379.43 km (95% CI: −502.43, −256.42), and the mean difference in distance east (longitude) among these groups was 225.60 km (95% CI: 154.42, 296.80). When we adjusted these differences for sunlight exposure, they both attenuated markedly to −95.18 km (95% CI: −178.34, −12.01) for the association with distance north (latitude) and 108.76 km (95% CI: 47.85, 169.67) for the association with distance east (longitude). The mean difference in sunshine (as percent of total daylight) comparing non-persistence (C) allele carriers with non-carriers was 0.48 (95% CI: 0.33, 0.64). After adjustment for latitude and longitude, this attenuated to 0.13 (95% CI: 0.02, 0.25).
Many of the characteristics examined in this study are related to geography (see Supplementary web Table 2). Given the geographic differences in allele frequency, together with geographical variation in the characteristics we have examined, we repeated all of the analyses presented in Table 2 with additional adjustment for latitude and longitude of birth. Supplementary web Table 4 (see journal website) gives these adjusted results. The association of non-persistence (C) allele homozygosity or heterozgosity and HDLc remained unchanged with additional adjustment for latitude and longitude of place of birth; 0.04 mmol/l (0.01, 0.07), P=0.01. The association of the non-persistence (C) allele homozygosity and self-rated poor or fair health attenuated from an odds ratio of 1.38 (1.04, 1.84) to one of 1.19 (0.87, 1.64) with adjustment for latitude and longitude of place of birth.
Discussion
Prevalence and geographical distribution
In a representative sample of white British women aged 60–79 years, we found a prevalence of the genotype associated with lactase non-persistence of 6.8%; among the 96% of these women who were born in Britain, the prevalence was 6.1%. The prevalence of phenotypically defined lactase non-persistence in participants described as ‘British born' or ‘white' has been estimated as ranging from 3 to 7%.18, 19, 20 Until recently, no large-scale studies with genotype data have been reported from Britain, but two small studies reported prevalences of 5.4 and 9.0% of non-persistence (C) allele homozygosity.21, 22 The latter study recruited participants from the south of England, where the prevalence of non-persistence (C) allele homozygosity in our study was higher than in the north of Britain. There was no evidence of an age difference by genotype, and there was no strong evidence of departure from the Hardy–Weinberg equilibrium either at younger or older ages, suggesting that in this population genotype is not related to survival.
The Wellcome Trust Case Control Consortium used the availability of both genomewide genotypic data and geographic information to assess evidence of unequal geographical distribution of genetic markers.23 This work noted a very small proportion (less than 1%) of loci that showed evidence for the difference in allele frequency by geographic region, the most marked of these being that of rs1042712, a marker proximal to the MCM6 locus (r2 with rs4988235=0.24; D′: 0.88; derived from the HAPMAP phase 2 CEU samples). This work did not unite this variation with patterns of phenotypic difference that might account for these differences.
The prevalence of lactase persistence shows strong associations with latitude, generally increasing with latitudes greater than 25° north or south.5 This association has usually been shown between countries rather than within countries. Here, we demonstrate that such a gradient exists within Britain; individuals with the lactase non-persistence (C) allele were more likely to have been born in the most southeasterly areas. These findings are supported by the aforementioned findings from the Wellcome Trust Case Control consortium. In this study, we show that this gradient depends on latitude of place of birth rather than on place of residence, suggesting that it reflects prevalence differences between peoples who have lived in different latitudes within Britain for many generations. These graded and substantial differences in prevalence of a genetic variant for a population all born in the United Kingdom and identified as being ‘white' demonstrate that potentially important levels of population substructure of genetic variants do exist within an apparently homogeneous population. The finding of an association between homozygosity for the lactase non-persistence allele and self-reported poor or fair health, which attenuated substantially on adjustment for latitude and longitude, further demonstrates that apparent associations with health outcomes can be generated by such stratification. Our findings are consistent with those of a recent study of an apparently homogeneous European American population, all of whom had described themselves as ‘white' or ‘Caucasian' and all of whom had grandparents born in either the United States or Europe.24 In that study, an apparent strong association between the LCT variant examined here and height was shown to be largely explained by population stratification. However, lactase persistence – a trait for which there is evidence of strong and recent selection6, 7 – may be an exception to a general rule of low levels of population stratification.
Lactase persistence, drinking milk and bone health
Between populations, there is a very strong association between the prevalence of lactase persistence and drinking milk.5 However, within populations, the association is much less strong. We found no strong association of milk consumption with lactase persistence, although our data on this issue were crude, consisting of a comparison of never drinking milk with drinking any amount, as opposed to a more graded assessment of amount consumed. Findings from previous studies have been mixed.25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 The power of many previous studies has been low because of small sample sizes given the time-consuming nature of phenotypic testing for lactase non-persistence. Lactase non-persistence tends to influence the amount of milk that can be tolerated rather than relating to being unable to drink milk at all, therefore our lack of data on the quantity of milk consumption is an important limitation in examining this issue.
There has been considerable discussion of the degree to which lactase non-persistence leads to ill-health. Recently, Matthews et al36 have demonstrated that lactase non-persistence variant homozygosity is associated with higher rates of many symptoms of ill-health in an UK population. We did not have questions to examine the symptoms for which strongest influences were found in that study – abdominal pain, gut distension, flatulence, diarrhoea, headache and generalised tiredness and pain36 – but questions on depression, asthma, gastric ulcer and nonspecific chest pain showed no strong association with genotype (data not shown). Our finding of a higher prevalence of poor or fair self-rated health appears to be generated at least, in part, by population substructure (as discussed above). Given the large number of possible statistical contrasts, any weak associations with genotype – for example, adult social class – need to be treated with considerable caution. The work by Matthews et al suggests that those who are lactase non-persistent but continue to drink milk are the ones most likely to report symptoms, but we found no difference in the effect of genotype on our general health rating when we stratified whether the woman drank milk or not. However, this analysis would be better performed taking into account amount of milk drunk, and we do not have data on this. Several studies have suggested associations between milk consumption and vascular and metabolic traits,37, 38, 39, 40, 41 and we have previously reported that women in this study who reported never drinking milk had lower levels of homoeostasis model assessment insulin resistance.42
We found no evidence that the LCT variant examined here was associated with insulin resistance and it was largely unrelated to vascular and metabolic traits. However, the lack of association with milk consumption in this population means that LCT cannot be used as an instrument for examining the causal effect of milk consumption on these outcomes. The association between possession of a C (lactase non-persistence) allele and HDLc remained even with the adjustment for latitude and longitude of birth, and hence may not be fully explained by population stratification. However, we have undertaken a number of statistical tests in this study, and replication of this finding would be required before one could consider this as anything other than a chance association.
Implications for the origin of lactase persistence
As a trait, lactase persistence shows strong evidence of selection, and there has been considerable discussion regarding the basis of this. The genetic haplotype associated with lactase persistence of which one perfect tag was investigated here also shows molecular evidence of selection, as does the variant recently identified in African populations that underlies a second origin of the trait.6, 7 There are several non-exclusive hypotheses regarding the transition from presumed near-universal lactase non-persistence in original Homo sapiens to high prevalences of lactase persistence in many populations today. They are all predicated on the notion that lactase persistence has evolved in parallel with the use of milk-producing animals in animal husbandry and the resultant potential for there to be large supplies of milk. Indeed, it has been shown that diversity in milk protein genetic variants among cattle is distributed across Europe in parallel to both current day lactase persistence and the Neolithic distribution of European cattle pastoralist societies.43
The two current main hypotheses regarding the origin of lactase persistence can be summarised as (1) the ‘culture historical' hypothesis,44, 45, 46 which concentrates on the general nutritional (and survival) advantage of milk consumption in populations that have milk availability, do not process milk into low-lactose foods such as cheese and are subject to other (non-dairy) dietary stresses and (2) the calcium absorption hypothesis,5, 47 which considers the ability to use milk as of particular importance for high-latitude populations with low ultraviolet light exposure who are thus subject to potential vitamin D deficiency and poor calcium absorption and for whom the calcium absorption-stimulating effect of lactose would increase fitness. There are other less formally articulated hypotheses that can be identified in the literature, which are as follows:5, 48, 49 (1) a reduced diarrhoeal disease mortality hypothesis that considers that, in populations that have become high consumers of milk, this consumption will increase risk of diarrhoeal disease in individuals who are not lactase persistent and thus select for lactase persistence; (2) an auxiliary water/electrolyte hypothesis specific to the aberrant high-lactase persistence populations in Africa that considers that, in arid regions with animal husbandry practices allowing access to milk, the ability to use milk has a selective advantage through the provision of water and electrolytes; (3) the enhanced fertility by early weaning hypothesis that postulates that lactase persistence leads to earlier weaning and that earlier cessation of breastfeeding reduces the infertile period following each birth; and finally, (4) that the gradients observed could be due to simple genetic drift.
It is possible that dairy farming became more strongly established at an earlier time in the north of Britain, although evidence for this is weak.50 When we adjusted the latitude association with hours of sunlight at the area of birth, we found marked attenuation of the latitude associations. In areas with lower sunlight exposure (and hence vitamin D levels), lactase persistence could provide a means of enhancing calcium absorption. However, the strong association of hours of sunlight with latitude renders statistical separation of these two influences difficult.
The reduced diarrhoeal disease hypothesis is dependent on the lack of a strong link between lactase persistence and milk consumption in cultures with high milk consumption and in this sense receives support from our findings. However, the consequences of prolonged childhood diarrhoeal disease that might be expected in survivors – shorter body height, leg length and perhaps higher blood pressure51, 52 – were not seen in our data (Table 2). Our study does not of course address the auxiliary water/electrolyte hypothesis. Finally, even if the enhanced fertility hypothesis applied to historical populations, it is unlikely to be reflected in recent fertility patterns in the United Kingdom, where total offspring number will not depend on periods of breastfeeding-induced fertility reductions. In our data, there was no association between genotype and parity (Table 2).
Further genotype-based studies across disparate populations and relating genotype to latitude, sunlight exposure, milk consumption, bone health, calcium absorption and diarrhoeal disease history could help progress understanding. For example, the finding that lactase–persistence-associated alleles are in different haplotypes in African as compared with European populations53 demonstrates a separate origin in these two continents and may provide some support for the auxiliary water/electrolyte intake hypothesis for high lactase persistence populations in Africa.
Implications for population stratification
Our finding of gradients in lactase persistence across Britain could relate to population origin (eg, a relatively greater contribution of high lactase persistence Norsemen in the north and of lower lactase persistence Germanic Anglo Saxons and Normans in the south50) or to differential strength of selection. Whatever the origins of these patterns, they demonstrate that common genetic variants may show considerable differences in prevalence between subgroups within apparently homogenous populations and thus that population substructure could generate bias in both estimates of precision of genetic variant – outcome associations and in the strength of these associations. A similar gradient in allele frequency within a country has recently been demonstrated within Italy.54 Lactase persistence would clearly be a useful variant for detecting population substructure and for use in statistical approaches for controlling the effects of such stratification.55 A recent study of European American populations has also identified this variant as one that is highly sensitive to population substructure24 and a subsequent paper using the same sample proposed a new, computationally simple method for correcting population stratification.56
Conclusions
Among a population of British-born women identified as being white, we show that there is a considerable population stratification for a common genetic variant, to a level that could produce spurious findings in genetic association studies. The lactase persistence variant could be useful in exploring the existence of, and then statistically controlling for, population stratification in genetic association studies. Lactase persistence is a classic genetic polymorphism identifiable through its phenotypic effect and as such has been included in the banks of genetic polymorphisms utilised for studying the origin and distribution of human genes.57, 58 Other such polymorphisms, some of which, like lactase persistence, can now be easily studied as SNPs, could also provide valuable data on hidden population subdivision.
Acknowledgments
The British Women's Heart and Health Study is funded by the UK Department of Health Policy Research Programme and the British Heart Foundation. DAL is funded by a UK Department of Health Career Scientist Award and NT by a UK Medical Research Council Studentship.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Disclosure
The views expressed in this manuscript are those of the authors and not necessarily those of the Department of Health, British Heart Foundation or Medical Research Council.
Supplementary Material
References
- Villako K, Maaroos H. Clinical picture of hypolactasia and lactose intolerance. Scand J Gastroenterol. 1994;29 Suppl 202:36–54. doi: 10.3109/00365529409091743. [DOI] [PubMed] [Google Scholar]
- Swallow DM, Hollox EJ.The genetic polymorphism of intestinal lactase activity in adult humansin Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic and Molecular Basis of Inherited Disease New York: McGraw-Hill; 20001651–1662. [Google Scholar]
- Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Jarvela I. Transcriptional regulation of the lactase–phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–652. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odling-Smee FJ, Laland KN, Feldman MW. Niche Construction: The Neglected Process in Evolution. Oxford: Princeton University Press; 2003. [Google Scholar]
- Durham WH.Cultural mediation: the evolution of adult lactose absorptionin Durham WH (ed):: Co-evolution: Genes, Culture and Human Diversity Stanford: Stanford University Press; 1991 [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–39. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- Rasinperä H, Savilahti E, Enattah NS, et al. A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut. 2004;53:1571–1576. doi: 10.1136/gut.2004.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter M, Hollox E, Harvey CB, et al. The causal element for the lactase persistence/non persistence polymorphism is located in a 1Mb region of linkage disequilibrium in Europeans. Ann Hum Genet. 2003;67:298–311. doi: 10.1046/j.1469-1809.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomark Prev. 2002;11:513–520. [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Wagging the dog? Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Witte JS. Point: population stratification: a problem for case–control studies of candidate–gene associations. Cancer Epidemiol Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–392. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Bedford C, Taylor M, Ebrahim S. Geographic variation in cardiovascular disease, risk factors and their control in older women: British Women's Heart and Health Study. J Epidemiol Commun Health. 2003;57:134–140. doi: 10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Ebrahim S, Davey Smith G. Socioeconomic position in childhood and adulthood and insulin resistance: cross sectional survey using data from the British women's heart and health study. BMJ. 2002;325:805–807. doi: 10.1136/bmj.325.7368.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MW, Povey S, Swallow D. Lactase polymorphism in adult British natives: estimating allele frequencies by enzyme assays in autopsy samples. Am J Hum Genet. 1982;34:650–657. [PMC free article] [PubMed] [Google Scholar]
- Ferguson A, Macdonald DM, brydon WG. Prevalence of lactase deficiency in British adults. Gut. 1984;25:163–167. doi: 10.1136/gut.25.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal TH, Wood GM, Lewis KO, Leek JP, Cooper BT. Prevalence of primary lactase deficiency in adult residents of west Birmingham. BMJ. 1993;306:1303. doi: 10.1136/bmj.306.6888.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasinperä H, Forsblom C, Enattah NS, et al. The C/C=13910 genotype of adult-type hypolactasia is associated with an increased risk of colorectal cancer in the Finnish population. Gut. 2005;54:643–647. doi: 10.1136/gut.2004.055939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow DM. Genetics of lactase persistence and lactose intolerance. Annu Rev Genet. 2003;37:197–219. doi: 10.1146/annurev.genet.37.110801.143820. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Newcomer AD, Thomas PJ, McGill DB, Hofmann AF. Lactase deficiency: a common genetic trait of the American Indian. Gastroenterology. 1977;72:234–237. [PubMed] [Google Scholar]
- Hohkanen R, Pulkkinen P, Jarvinen R, et al. Does lactose intolerance predispose to low bone density? A population-based study of permenopausal Finnish women. Bone. 1996;19:23–28. doi: 10.1016/8756-3282(96)00107-x. [DOI] [PubMed] [Google Scholar]
- Obermayer-Pietsch BM, Bonelli CM, Walter DE, et al. Genetic predisposition to adult lactose intolerance and relation to diet, bone density, and bone fractures. J Bone Miner Res. 2004;19:42–47. doi: 10.1359/JBMR.0301207. [DOI] [PubMed] [Google Scholar]
- Horowitz M, Wishart J, Mundy L, Nordin BEC. Lactose and calcium absorption in postmenopausal osteoporosis. Arch Intern Med. 1987;147:534–536. [PubMed] [Google Scholar]
- Newcomer AD, Hodgson SF, Douglas MD, Thomas PJ. Lactase deficiency: prevalence in osteoporosis. Ann Intern Med. 1978;89:218–220. doi: 10.7326/0003-4819-89-2-218. [DOI] [PubMed] [Google Scholar]
- Rasinperä H, Savilahti E, Enattah NS, et al. A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut. 2004;53:1571–1576. doi: 10.1136/gut.2004.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguet P, Faille I, Destrebecq L, Devogelaer J-P, Nagant de Deuxchaisnes C. Lactose intolerance, calcium intake, and osteopenia. Lancet. 1991;338:1156–1157. doi: 10.1016/0140-6736(91)92024-v. [DOI] [PubMed] [Google Scholar]
- Obermayer-Pietsch BM, Gugatschka M, Reitter S, et al. Adult-type hypolactasia and calcium availability: decreased calcium intake or impaired calcium absorption. Osteoporos Int. 2007;18:445–451. doi: 10.1007/s00198-006-0251-6. [DOI] [PubMed] [Google Scholar]
- Stephenson LS, Latham MC. Lactose intolerance and milk consumption: the relation of tolerance to symptoms. Am J Clin Nutr. 1974;27:296–303. doi: 10.1093/ajcn/27.3.296. [DOI] [PubMed] [Google Scholar]
- Corazza GR, Benati G, Di Sario A, et al. Lactose intolerance and bone mass in postmenopausal Italian women. Br J Nutr. 1995;73:479–487. doi: 10.1079/bjn19950050. [DOI] [PubMed] [Google Scholar]
- Harma M, Alhava E. Is lactose malabsorption a risk factor in fractures of the elderly. Annales Chirurgiae et Gynaecologiae. 1988;77:180–183. [PubMed] [Google Scholar]
- Matthews SB, Waud JP, Roberts AG, Campbell AK. Systematic lactose intolerance: a new perspective on an old problem. Postgrad Med J. 2005;81:167–173. doi: 10.1136/pgmj.2004.025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JJ. Milk and coronary heart disease mortality. J Epidemiol Community Health. 2002;56:319. doi: 10.1136/jech.56.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MA, Jacobs DR, Jr, van Horn L, Slattery ML, Kartashov AI, Ludwig DS. Dairy consumption, obesity, and the insulin resistance syndrome in young adults: the CARDIA Study. JAMA. 2002;287:2081–2089. doi: 10.1001/jama.287.16.2081. [DOI] [PubMed] [Google Scholar]
- Mennen LI, Lafay L, Peskena EJM, Novak M, Lepinay P, Balkau B. Possible protective effect of bread and dairy products on the risk of metabolic syndrome. Nutr Res. 2000;20:335–347. [Google Scholar]
- Massey LK. Dairy food consumption, blood pressure and stroke. J Nutr. 2001;131:1875–1878. doi: 10.1093/jn/131.7.1875. [DOI] [PubMed] [Google Scholar]
- Miller ER, Appel LJ, Riaby TH.Effects of dietary patterns on measures of lipid peroxidation Circulation 2000102852–857.10952952 [Google Scholar]
- Lawlor DA, Ebrahim S, Timpson N, Davey Smith G. Avoiding milk is associated with reduced risk of insulin resistance and the metabolic syndrome: findings from the British Women's Heart and Health Study. Diabet Med. 2005;22:808–811. doi: 10.1111/j.1464-5491.2005.01537.x. [DOI] [PubMed] [Google Scholar]
- Beja-Pereira A, Luikart G, England PR, et al. Gene–culture coevolution between cattle milk protein genes and human lactase genes. Nat Genet. 2003;35:311–313. doi: 10.1038/ng1263. [DOI] [PubMed] [Google Scholar]
- Simoons FJ. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations. II. A culture historical hypothesis. Am J Dig Dis. 1970;15:695–710. doi: 10.1007/BF02235991. [DOI] [PubMed] [Google Scholar]
- McCracken RD. Lactase deficiency: an example of dietary evolution. Curr Anthropol. 1971;12:479–517. [Google Scholar]
- Simoons FJ. Persistence of lactase activity among Northern Europeans: a weighing of evidence for the calcium absorption hypothesis. Ecol Food Nutr. 2001;40:397–469. [Google Scholar]
- Flatz G, Rotthauwe HW. Lactose nutrition and natural selection. Lancet. 1973;2:76–77. doi: 10.1016/s0140-6736(73)93267-4. [DOI] [PubMed] [Google Scholar]
- Cook GC. Did persistence of intestinal lactase into adult life originate on the Arabian Peninsula. Man. 1978;13:418–427. [Google Scholar]
- Lieberman M, Lieberman D. Lactase deficiency: a genetic mechanism which regulates the time of weaning. Am Nat. 1978;112:625–639. [Google Scholar]
- Miles D. The Tribes of Britain. London: Weidenfeld and Nicolson; 2005. [Google Scholar]
- Davey Smith G, Leary S, Ness A. Could dehydration in infancy lead to high blood pressure. J Epidemiol Community Health. 2006;60:142–143. doi: 10.1136/jech.2005.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Davey Smith G, Mithcell R, Ebrahim S. Adult blood pressure and climate conditions in infancy: a test of the hypothesis that dehydration in infancy is associated with higher adult blood pressure. Am J Epidemiol. 2006;163:608–614. doi: 10.1093/aje/kwj085. [DOI] [PubMed] [Google Scholar]
- Mulcare CA, Weale ME, Jones AL, et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C–13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet. 2004;74:1102–1110. doi: 10.1086/421050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdote C, Guarrera S, Davey Smith G, et al. Lactase persistence and bitter taste response: instrumental variables and Mendelian randomization in epidemiologic studies of dietary factors and cancer risk. Am J Epidemiol. 2007;166:576–581. doi: 10.1093/aje/kwm113. [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Allen AS, Satten GA. A simple and improved correction for population stratification in case–control studies. Am J Hum Genet. 2007;80:921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury AK, Nei M. Human Polymorphic Genes. World Distribution. Oxford: Oxford University Press; 1998. [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NH: Princeton University Press; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.