Abstract
Fibrodysplasia ossificans progressiva (FOP, MIM 135100) is a rare genetic disorder characterized by congenital great toe malformations and progressive heterotopic ossification transforming skeletal muscles and connective tissues to bone following a well-defined anatomic pattern of progression. Recently, FOP has been associated with a specific mutation of ACVR1, the gene coding for a bone morphogenetic protein type I receptor. The identification of ACVR1 as the causative gene for FOP now allows the genetic screening of FOP patients to identify the frequency of the identified recurrent ACVR1 mutation and to investigate genetic variability that may be associated with this severely debilitating disease. We report the screening for mutations in the ACVR1 gene carried out in a cohort of 17 Italian patients. Fifteen of these displayed the previously described c.617G>A mutation, leading to the R206H substitution in the GS domain of the ACVR1 receptor. In two patients, we found a novel mutation c.774G>C, leading to the R258S substitution in the kinase domain of the ACVR1 receptor. In the three-dimensional model of protein structure, R258 maps in close proximity to the GS domain, a key regulator of ACVR1 activity, where R206 is located. The GS domain is known to bind the regulatory protein FKBP12 and to undergo multiple phosphorylation events that trigger a signaling cascade inside the cell. The novel amino-acid substitution is predicted to influence either the conformation/stability of the GS region or the binding affinity with FKBP12, resulting in a less stringent inhibitory control on the ACVR1 kinase activity.
Keywords: FOP, ACVR1, heterotopic ossification, BMPs
Introduction
Fibrodysplasia ossificans progressiva (FOP, MIM 135100) is a rare genetic disorder characterized by congenital malformations of the great toes and progressive heterotopic ossification causing the replacement of skeletal muscle and soft connective tissues with bone following a well-defined anatomic pattern.1, 2, 3 FOP is the most severe and disabling disorder of heterotopic ossification in humans and leads to the formation of a second skeleton. Heterotopic ossification begins in childhood either spontaneously or following soft tissue injury and progresses in adulthood with typical episodic flare-ups and remissions.1, 2, 3 The severe disability of FOP results in low reproductive fitness. The condition is usually sporadic because of the occurrence of a spontaneous mutation. However, some familial cases have been described with an autosomal dominant pattern of inheritance and variable expression.4, 5 Genome-wide linkage analysis in these families followed by candidate gene screening identified ACVR1 as the gene responsible for FOP. All familial and sporadic cases of classic FOP reported to date are heterozygous for the same mutation, c.617G>A, leading to the amino-acid substitution, R206H.4, 6, 7 A series of patients with phenotypic and genotypic variants of FOP was recently described.4, 6, 7, 8 An additional patient with an FOP variant was recently identified with a de novo ACVR1 mutation, G356R, associated with the disease.9
The ACVR1 gene encodes the activin A type I receptor (also known as activin receptor-like kinase 2, ALK2), a receptor for bone morphogenetic proteins (BMPs).10 ACVR1 is a type I serine/threonine receptor kinase belonging to the transforming growth factor-β receptor (TGFBR1) family, composed of seven receptors (ALKs 1–7) (for a review, see Graham and Peng11), which, together with type II receptors, form a heterotetrameric complex at the cell membrane.12, 13, 14 Both receptor types have an extracellular ligand-binding domain, a single transmembrane domain and a cytoplasmic serine/threonine kinase domain. Type I receptors contain an additional regulatory domain that is not present in type II receptors. This region, known as the GS domain for its conserved glycine–serine-rich sequence, is phosphorylated by type II receptors to trigger the signaling cascade inside the cell following receptor-binding by ligand.13, 14, 15 Downstream events are mediated by phosphorylation of the regulatory SMAD proteins (R-SMADs), which bind SMAD 4. Then, the complex translocates to the nucleus where it modulates the expression of specific target genes.16 By contrast, inhibitory SMADs, such as SMAD 6 and 7, are involved in the ligand-dependent termination of signaling.16
The conserved GS domain plays an important regulatory role for type I receptors. It is well documented that this region physically interacts with the FK506-binding protein 12 (FKBP12) in TGFBR1,17, 18 and more recently, this has been confirmed also for the activin type I receptor, ALK4.19 FKBP12 is a cytoplasmic protein that binds to immunosuppressant drugs, such as FK506 and rapamycin with high affinity. Besides the immunosuppressive effect, an influence on bone metabolism has recently been observed for FK506. In particular, when administered locally or in vitro in combination with BMPs, FK506 can induce osteogenesis.20 Recent evidence shows that FKBP12 binding to the GS domain of type 1 BMP receptors helps to maintain the receptor inactivation, preventing leaky signaling in the absence of ligand. Because of high-affinity interaction, FK506 can displace FKBP12 from the receptor, which is then able to transduce downstream signaling and to promote osteogenesis.20 Additionally, FKBP12 plays a second functional role on the GS domain. Recent studies on the ALK4 receptor demonstrate that, upon ligand stimulation, FKBP12 transiently dissociates from the activated receptor. Rebinding to the ALK4 GS region, a few hours later, mediates the recruitment of inhibitory SMAD7 and SMURF1 (smad ubiquitin regulatory factor 1), an E3 ubiquitine ligase. The resulting receptor ubiquitination causes the termination of the intracellular signaling.19 FKBP12 is a member of a protein family composed, in the human genome, by at least 15 distinct homologous genes.21 To date, experimental evidence of a direct interaction of ACVR1 with FKBP12 is still lacking. It is, however, likely that either FKBP12 or one of its paralogues has a regulatory role in ACVR1 activity.
Although the three-dimensional structure of ACVR1 has not been determined, the crystal structure of the homologous TGFBR1 in complex with FKBP124, 22 can be used to build, by homology, a three-dimensional model of the cytoplasmic portion of ACVR1. The good reliability of the model is determined by the high degree of sequence similarity between ACVR1 and TGFBR1 (66% identical residues) and the lack of insertions or deletions in the pairwise sequence alignment. Using such model, it was possible to show that the basic arginine residue, R206, at the end of the ACVR1 GS domain of the protein, conserved across mammals and mutated in FOP patients, forms a salt bridge with an invariant aspartate residue (D269 in ACVR1) that links the GS domain to the L45 loop, a SMAD specificity site.4, 23 Therefore, it has been hypothesized that the specific substitution of arginine 206 with histidine could introduce a pH-sensitive switch and induce a conformational change at decreased intracellular pH, thus leading to ligand-independent activation of the receptor.23
In this work, we describe the mutational analysis for the ACVR1 gene carried out on 17 Italian FOP patients. We found the recurrent R206H mutation in 15 cases and in two patients, we identified the new mutation, c.774G>C, leading to the substitution of R258S in the kinase domain. We also performed an in silico analysis for both the previously described R206H and the novel R258S mutation to evaluate their effects on the receptor function. For this purpose, we also generated the three-dimensional model for the cytoplasmic domain of the R258S ACVR1 mutant.
Materials and methods
Patients and diagnostic criteria
Clinical diagnosis of FOP was based on two main criteria: malformation of the great toes and presence of heterotopic ossification with a specific anatomic and temporal pattern. In addition to the congenital hallux malformation, patients with FOP could have other less penetrant skeletal malformations, such as thumb malformations, orthotopic fusions of posterior elements of the cervical spine, short and broad femoral neck and proximal tibial osteochondromas. Heterotopic ossification is preceded by the formation of soft tissue, tumor-like lesions appearing between 2 and 5 years of life. Most of these lesions then give rise to ectopic bone neoformation through an endochondral process. Following these criteria, 17 Italian patients with a clinical diagnosis of FOP were referred to our Laboratory and considered eligible for the mutational analysis of the ACVR1 gene. The malformation of the great toe was absent in one case, FOP12 patient, for which clinical diagnosis of FOP was based on the course and localization of heterotopic ossification. The heterotopic ossification was absent in one case, FOP15, for which early clinical diagnosis of FOP was based on malformation of great toe, fusion of posterior elements of the cervical spine and presence of a proximal tibial osteochondroma.24, 25, 26
Peripheral blood samples were obtained from patients, in some cases from their parents, and from control individuals with an informed consent.
DNA extraction, PCR and sequencing reactions
Genomic DNA was extracted from peripheral blood samples with the Puregene Blood Kit (Gentra Systems Inc., Minneapolis, MN, USA; www.gentra.com) according to the manufacturer's procedure. Oligonucleotides and conditions used to amplify ACVR1 coding sequence were already described in Shore et al.4 PCR reactions were carried out in a total volume of 20 μl containing 1 × PCR buffer (Applied Biosystems, Foster City, CA, USA; www.appliedbiosystems.com), 200 μ dNTPs mix, 1.5 m MgCl2, 10 pmoles of each oligonucleotide, 1 U of AmpliTaq Gold Polymerase (Applied Biosystems).
PCR products were checked by standard agarose gel electrophoresis and purified by Exo/SAP-IT (USB Europe GmbH, Staufen, Germany; www.usbweb.com) digestion: briefly, 5 μl of PCR product were incubated with 2 μl of the Exo/SAP-IT reagent at 37°C for 30 min, inactivated at 80°C for 15 min and then used for direct sequencing. Sequencing reactions were set up with a Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems) according to the provided protocol and run on a 3130xl Genetic Analyzer (Applied Biosytems).
cDNA analysis
Total RNA was extracted from lymphocytes with the RNeasy Mini Kit (Qiagen, GmbH, Hilden, Germany; www1.qiagen.com) according to the manufacturer's protocol. In all, 1 μg of RNA was used for retrotranscription by using the Advantage RT-for-PCR kit (Becton Dickinson, BD Biosciences Clontech, San José, CA, USA; www.bdbioscience.com). Briefly, first strand cDNA was synthesized by oligo(dT) priming for 1 h at 42°C, 5′ at 94°C and diluted 1:5 for PCR reactions. cDNA synthesis was checked by PCR with oligonucleotides for human G3PDH included in the Advantage RT-for-PCR kit. Amplification of specific ACVR1 cDNA was obtained by using a forward primer mapping in the third untranslated exon combined with a reverse located in the sixth protein coding exon (F- AGTGAGAGAAGCTCTGAACG and R- CGAAGGCAGCTAACTGTATC), according to the NM_001105 RefSeq (www.ncbi.nlm.nih.gov/Entrez). The specific 1001 bp product was then subcloned using the TOPO-TA cloning Kit (Invitrogen; Carlsbad, CA, USA; www.invitrogen.com) to separate the two different alleles. Twelve colonies were selected and grown overnight in Luria–Bertani medium. Plasmids were then recovered with the IllustraTM plasmidPrep Mini Spin Kit (GE Healthcare UK Limited Little Chalfont, Buckinghamshire, UK; www.gehealthcare.com) according to the provided protocol. Plasmid DNA was sequenced by using the M13 forward and reverse vector primers included in the TOPO-TA cloning kit (Invitrogen) and the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems) as described previously.
Sequence analysis and bioinformatics tools
Sequence alignments were obtained with the ClustalX software.27 that was freely downloaded from the Bioinformatics support group of the Conway Institute UCD (Dublin, Ireland) site at http://www.clustal.org. The single nucleotide polymorphism database (dbSNP) is available through the NCBI home page at http://www.ncbi.nlm.nih.gov/SNP. PolyPhen (polymorphism phenotyping) and SIFT (sorting intolerant from tolerant) are bioinformatics tools that allow to predict the effect of an amino-acid substitution on the structure and function of a protein using physical and comparative methods. They are freely available at the following sites: http://genetics.bwh.harvard.edu/pph/ (PolyPhen) and http://blocks.fhcrc.org/sift/SIFT.html (SIFT), respectively.28, 29, 30, 31
Molecular modeling
The three-dimensional models of the cytoplasmic domain of ACVR1 and of the R258S ACVR1 mutant were built by homology using the crystallographic structure of the type I TGF-β receptor in complex with the immunophilin inhibitory protein FKBP12 (PDB code, 1PY5)22 as template structure. The cytoplasmic domain of ACVR1 displays substantial sequence similarity (66% identical residues along the amino-acid stretch 170–500) with the same domain of TGFBR1. In the aligned stretch, no insertions or deletions are found. The two models were built and energy minimized with the SwissModel program suite32 and visually analyzed by using the O program suite.33 Although the calculated model of the cytoplasmic domain of ACVR1 cannot replace a direct experimental structural investigation, the calculated theoretical models should be considered reliable in consideration of the high level of sequence similarity and of the lack of insertions and deletions. However, the side chain conformation, especially that of mutated residues, could be imprecise and should be considered with caution. This is also because of the limitations of the energy minimization procedure adopted by SwissModel, which does not use molecular dynamics or simulated annealing protocols, and the lack of reliable molecular modeling procedures.
Results
We have examined 17 patients with a clinical diagnosis of FOP. All the patients are Italian with no family history for FOP. First, we looked for the presence of the common FOP mutation, the c.617G>A substitution in the fourth protein coding exon. As reported in Table 1, 15 patients were heterozygous for the recurrent substitution (Figure 1a), whereas FOP12 and FOP17 were negative. The parents of only three patients with the c.617G>A substitution could be examined for the presence/absence of the mutation, thus demonstrating the de novo origin of the mutation.
Table 1. Summary of the patient's clinical features.
| Patient | Age | Sex | Age ossification onset | Great toe malformation | Mutation | Other |
|---|---|---|---|---|---|---|
| FOP1 | 32 years | M | NA | Yes | c.617G>A, nd | |
| FOP2 | 4 years | M | 15 months | Yes | c.617G>A, de novo | Hydrocephalus secondary to a posterior bulbar expansive lesion of unknown origin (surgical correction) |
| FOP3 | 9 years | F | 4 years | Yes | c.617G>A, de novo | |
| FOP4 | 4 years | F | 20 months | Yes | c.617G>A, de novo | |
| FOP5 | NA | M | 8 years | Yes | c.617G>A, nd | |
| FOP6 | 28 years | F | 6 years | Yes | c.617G>A, nd | |
| FOP7 | 27 years | F | 11 years | Yes | c.617G>A, nd | |
| FOP8 | 49 years | F | 11 years | Yes | c.617G>A, nd | |
| FOP9 | 44 years | M | 3 years | Yes | c.617G>A, nd | |
| FOP10 | 35 years | M | 14 years | Yes | c.617G>A, nd | |
| FOP11 | 9 years | M | 6 months | Yes | c.617G>A, nd | |
| FOP12 | 21 years | F | 4 years | No | c.774G>C, de novo | c.44C>G dbSNP rs13406336, inherited |
| FOP13 | 48 years | M | 5 years | Yes | c.617G>A, nd | Episodic seizures |
| FOP14 | 36 years | M | 4 years | Yes | c.617G>A, nd | |
| FOP15 | 8 years | M | 6 years | Yes | c.617G>A, nd | |
| FOP16 | 16 years | F | NA | Yes | c.617G>A, nd | |
| FOP17 | 42 years | F | 14 years | Yes | c.774G>C, nd |
dbSNP, single nucleotide polymorphism database; NA, not available; nd, mutation origin not determined.
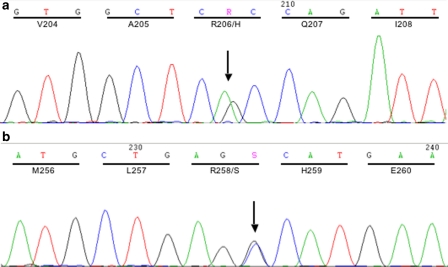
Figure 1.
Electropherograms of the two point mutations found in our case series: the c.617G>A substitution, recurrent in FOP (a), and the new c.774G>C transition (b) causing the R206H and R258S residue change, respectively.
In the two cases who were negative for the c.617G>A substitution (FOP12 and FOP17), we have analyzed the complete ACVR1 coding sequence by PCR amplification of genomic DNA and direct sequencing. In both patients, we found the novel ACVR1 mutation c.774G>C (Figure 1b) leading to the substitution, R258S, in the kinase domain of the protein. This nucleotide substitution was neither present in the healthy parents of FOP12 patient, nor in a panel of 104 control individuals. Moreover, it is not recorded in the dbSNP and has not been reported in the literature. The FOP12 patient carrying this new ACVR1 R258S variant had disease onset at the age of 4 years with painful swelling in the cervical vertebral region. Subsequently, the patient had a long remission period without additional clinical manifestation. At 18 years of age, the disease flared-up with progressive heterotopic ossification following the typical anatomic distribution. From a clinical point of view, the only striking peculiarity of this patient was the absence of great toe malformation. All other patients, including those with the recurrent mutation and FOP17 carrying the R258S variant, had the great toe malformation, although this presented rather mild in FOP17, and showed an extreme variability in the severity of the disease.
The FOP12 patient is also heterozygous for a c.44C>G substitution in exon 1, causing the A15G amino-acid change in a region predicted to encode the protein signal peptide. This variant was inherited from the healthy father and is reported in the dbSNP (rs13406336). cDNA analysis in the FOP12 patient showed that the R258S substitution occurred on the paternal allele, in cis configuration with the exon 1 variant. In our case series, the polymorphism was also found in a non-FOP patient, diagnosed as osteoma cutis, and was absent in a panel of 50 unaffected controls.
The newly identified mutation affected the R258 residue, which is highly conserved among species (Figure 2a), as well as among the members of the TGFBR1 family (Figure 2b). To assess the effect of the R258S substitution on protein structure and function, we have applied two freely available web-based services, PolyPhen and SIFT.29, 34 Both programs were developed to predict whether a specific amino-acid substitution is deleterious for protein function. The programs rely on structural information obtained with crystallographic or NMR methods, on other functional and structural characterizations and on primary sequence comparison, and are trained on a number of proteins and mutants having known mutation–phenotype relationships. If sufficient homology and structural data are available for a specific protein, both programs predict neutral or deleterious substitutions providing a score of significance for each prediction.28, 29, 30, 31 PolyPhen and SIFT analyses were conducted for R258S and for the R206H recurrent mutation to compare the results obtained for the new mutation. The results of this analysis are summarized in Table 2. Both substitutions affect residues, which are highly conserved through evolution, and both programs predict a deleterious effect on protein structure and function with highly significant scores (Table 2).
Figure 2.
The R258 residue, boxed in the figure, is highly conserved among species (a) and among the members of the ALK protein family (b). According to the ClustalX software (see Materials and methods), the asterisks underneath sequences in panel b indicate positions with a single, fully conserved residue.
Table 2. Bioinformatic analysis of the R206H and R258S substitutions.
| WT residue | Mutated residue | PolyPhen prediction | SIFTa prediction | |
|---|---|---|---|---|
| ACVR1 p.R206H | Positively charged/ hydrophilic | Positively charged/ hydrophilic | Probably damagingb; PSIC score 2.348c | Affects protein function; score 0.00 (M 3.04; S 52) |
| ACVR1 p.R258S | Positively charged/ hydrophilic | Neutral/polar/hydrophilic/ Possible phosphorylation | Probably damagingb; PSIC score 2.764c | Affects protein function; score 0.00 (M 3.07; S 29) |
PolyPhen and SIFT web services were queried to predict the effects on protein structure and function.
SIFT (sorting intolerant from tolerant) score is the normalized probability that the residue substitution is tolerated; score <0.05 indicates that the amino-acid change is predicted to affect the protein function.
PolyPhen results are empirically defined as follows: probably damaging – it is with high confidence likely to affect protein function or structure; possibly damaging – it is supposed to affect protein function or structure; benign – most likely lacking any phenotypic effect; unknown – the lack of data do not allow PolyPhen to make a prediction.
A PSIC score >2 indicates that the substitution is predicted to be damaging for protein structure or function and is only rarely or never observed in this position among different species
M represents the ‘median sequence information', a parameter measuring the diversity of the sequences used for prediction. S represents the number of sequences used for alignments at the position of the substitution. Sequences with gaps in the region are not considered.
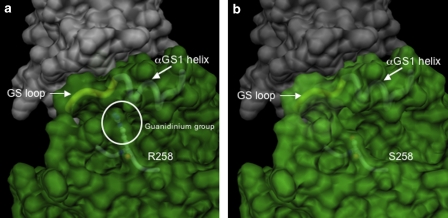
The three-dimensional model structure of the ACVR1 protein shows that both R206 and R258 are surface residues. Unlike the R206 residue, R258 is not part of the GS domain and does not appear to be involved in direct interaction with putative physiological ACVR1-binding proteins, such as FKPB12. However, the side chain of R258 is packed against the GS loop, and it provides a unique molecular side contact for the amino-acid stretch spanning amino-acid positions 185–190 (Figure 3a). The substitution of the arginine 258 side chain with the smaller serine side chain leaves the former stretch without side support (Figure 3b).
Figure 3.
Representation of the molecular surface of the ACVR1: FKPB12 complex in the proximity of the mutated site. (a) Wild-type ACVR1. On account of the strong similarity between the catalytic domain of TGFBR1 and ACVR1, the quaternary arrangement of the ACVR1: FKPB12 complex is predicted to be the same as that observed in the crystal structure of the TGFBR1: FKPB12 complex.22 The surfaces of the two proteins were calculated independently with the MSMS program suite32, 33 with a conventional probe radius of 1.4 Å. The residue, R258, is represented in ball-and-stick, visible under the semitransparent surface, together with the main chain in its close proximity. The main chain of the αGS1 helix and the GS loop are schematically represented and indicated, the circle encloses the guanidium group of the R258 residue. (b) R258S mutation. The shorter serine side chain is no longer in interaction with the GS region.
Discussion
Our cohort of 17 patients representing about 50% of the expected number of Italian FOP patients (based on the frequency of FOP or 1 per 2 million and an Italian population of 60 million), has shown that most patients (15/17) have classic FOP features and the recurrent heterozygous R206H mutation. Of the two patients carrying the newly identified R258S substitution, one showed an FOP variant phenotype, as described in Results, whereas the other showed a classic FOP phenotype. In the variant patient, the typical great toe malformation was absent and the diagnosis of FOP was made based on the course and localization of heterotopic ossification.
Our finding is in accordance with previous reports of ACVR1 protein kinase domain mutations in few cases of FOP defined as phenotypically variant,4, 6, 7, 8 and with a very recent published report by Furuya et al9 that described a patient affected with FOP showing a slow clinical course, sensorineural hearing loss and hypodactyly, which is associated with a novel mutation (G356D), affecting a conserved residue of the ACVR1 kinase domain. However, we underscore that the rare R258S mutation was carried by a patient showing no particular phenotypic variation and that, in our series of 17 cases, two were carrying this newly identified mutation, which is certainly de novo in one case and probably also in the second one, although we could not analyze the unaffected parents' DNA.
The FOP12 patient carrying the R258S substitution is also heterozygous for the A15G amino-acid change in the putative protein signal peptide and the two variants are in cis configuration. The polymorphism was inherited from the healthy father and is reported in the dbSNP (rs13406336). Assessing the population frequency of this variant, we found two conflicting data sets submitted to the HapMap Project Consortium and reported in the dbSNP. In one report, the G allele is virtually absent, whereas in a more recent submission, this allele is reported at a frequency of 0.5. The data obtained in our case series and in a panel of control individuals support that A15G is a rare ACVR1 variant. Its localization in the putative protein signal peptide suggests that it could affect the efficiency of receptor transportation to the cell membrane. However, the presence of the A15G variant in healthy individuals (evidenced by FOP12's father, controls and the record in the dbSNP) supports that this amino-acid variation is likely to be well tolerated with little, if any, role in FOP pathogenesis.
The newly described R258S substitution supports and extends the hypotheses on the functional effect of ACVR1 mutations in FOP. The three-dimensional structure of the ACVR1 protein shows that both R206 and R258 are surface residues, with the R206 residue belonging to the GS domain and the R258 to the kinase domain of the receptor. The R258 amino acid does not appear to be involved in direct interaction with putative physiological ACVR1-binding proteins, such as FKPB12 and its substitution is not expected to have immediate destabilizing effect on the conformation of the GS loop. Indeed, this region maintains unchanged interactions with the protein core below and with FKBP12 on the other side. However, the structural changes caused by multiple phosphorylations in the GS loop that are responsible for the activation of the kinase activity of ACVR1 are likely to be more easily achieved if the GS loop is less firmly anchored to the remaining part of the protein. This structural alteration may result in the decreased binding affinity for FKBP12 or in a direct change of the conformation of the regulatory αC helix. Both possibilities support a gain-of-function effect. Such hypotheses can also apply to the substitution reported by Furuya et al9 at the ACVR1 G356 amino acid, located in the protein core in a position contiguous to the αC helix. The replacement of glycine with aspartate, that has a bulkier and charged side chain, is likely to have a destabilizing effect on the αC helix, which is known to play a key role in the regulation of the kinase activity of TGFBR1.15, 22 Further in vitro experiments should address this important question.
The molecular characterization of ACVR1 sequence variation in a cohort that is likely to represent 50% of the Italian FOP population extends our understanding of genetic variation that causes FOP. Data of our case series confirm great variability in severity of the disease, including differences in age of onset, flare-up inducing stimuli, frequency and duration of flare-ups, rate of progression and different consequences on the quality of life of affected people, with some reaching a more severe degree of disability very early in childhood and others later in adulthood, as already reported.3, 4, 35 Interestingly, studies of monozygotic twins with FOP, all of them presenting with congenital toe malformations, but displaying variability in clinical phenotype evolution, suggest that environmental factors also can affect the phenotypic variability.36
Our findings add relevant information that contributes to identify the molecular and cellular mechanisms that cause FOP and for developing targets for therapeutic intervention.
Acknowledgments
We thank the patients and their families, and in particular we are grateful to the ‘FOP Italia Association' for their fundamental help and enthusiastic support to our work. We acknowledge the contribution of Drs Margherita Lerone, Maria Teresa Divizia, Maria Alpigiani, Maria Cristina Schiaffino and Professor Gianfranco Ferraccioli who referred patients. We also thank the ‘Cell line and DNA Bank from patients affected by Genetic Diseases' collection at the ‘Diagnosi Pre-Postnatale Malattie Metaboliche' Laboratory of G Gaslini Institute, Biobank funded by Telethon Grants (project no GTF04002) for providing specimens. We greatly acknowledge Drs Eileen Shore and Frederick Kaplan for invaluable help in discussion of our results and critical revision of the manuscript.
Financial contribution from the FOP Italia Association and from Fondazione CARIGE are greatly acknowledged.
References
- Kaplan FS, Glaser DL, Hebela N, Shore EM. Heterotopic ossification. J Am Acad Orthop Surg. 2004;12:116–125. doi: 10.5435/00124635-200403000-00007. [DOI] [PubMed] [Google Scholar]
- Kaplan FS, Galser DL, Shore EM, et al. The phenotype of fibrodysplasia ossificans progressiva. Clin Rev Bone Miner Metab. 2005;3:201–204. [Google Scholar]
- Pignolo RJ, Suda RK, Kaplan FS. The fibrodysplasia ossificans progressiva lesion. Clin Rev Bone Miner Metab. 2005;3:195–200. [Google Scholar]
- Shore EM, Xu M, Feldman GJ, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- Shore EM, Feldman G, Xu M, Kaplan FS. The genetics of fibrodysplasia ossificans progressiva. Clin Rev Bone Miner Metab. 2005;3:201–204. [Google Scholar]
- Lin GT, Chang HW, Liu CS, Huang PJ, Wang HC, Cheng YM. De novo 617G-A nucleotide mutation in the ACVR1 gene in a Taiwanese patient with fibrodysplasia ossificans progressiva. J Hum Genet. 2006;51:1083–1086. doi: 10.1007/s10038-006-0069-2. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Haga N, Takikawa K, Manabe N, Nishimura G, Ikegawa S. The ACVR1 617G>A mutation is also recurrent in three Japanese patients with fibrodysplasia ossificans progressiva. J Hum Genet. 2007;52:473–475. doi: 10.1007/s10038-007-0128-3. [DOI] [PubMed] [Google Scholar]
- Shore EM, Xu M, Connor JM, Kaplan FS.Twenty-Eighth Annual Meeting of the American Society for Bone and Mineral Research Philadelphia, Pennsylvania, USA: The American Society for Bone and Mineral Research; 2006. pp S75 [Google Scholar]
- Furuya H, Ikezoe K, Wang L, et al. A unique case of fibrodysplasia ossificans progressiva with an ACVR1 mutation, G356D, other than the common mutation (R206H) Am J Med Genet A. 2008;146A:459–463. doi: 10.1002/ajmg.a.32151. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Graham H, Peng C. Activin receptor-like kinases: structure, function and clinical implications. Endocr Metab Immune Disord Drug Targets. 2006;6:45–58. doi: 10.2174/187153006776056585. [DOI] [PubMed] [Google Scholar]
- Lin HY, Moustakas A. TGF-beta receptors: structure and function. Cell Mol Biol (Noisy-le-grand) 1994;40:337–349. [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Wang T, Li BY, Danielson PD, et al. The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell. 1996;86:435–444. doi: 10.1016/s0092-8674(00)80116-6. [DOI] [PubMed] [Google Scholar]
- Wang T, Donahoe PK. The immunophilin FKBP12: a molecular guardian of the TGF-beta family type I receptors. Front Biosci. 2004;9:619–631. doi: 10.2741/1095. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kurisaki A, Yamakawa N, Minakuchi K, Sugino H. FKBP12 functions as an adaptor of the Smad7-Smurf1 complex on activin type I receptor. J Mol Endocrinol. 2006;36:569–579. doi: 10.1677/jme.1.01966. [DOI] [PubMed] [Google Scholar]
- Kugimiya F, Yano F, Ohba S, et al. Mechanism of osteogenic induction by FK506 via BMP/Smad pathways. Biochem Biophys Res Commun. 2005;338:872–879. doi: 10.1016/j.bbrc.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Barik S. Immunophilins: for the love of proteins. Cell Mol Life Sci. 2006;63:2889–2900. doi: 10.1007/s00018-006-6215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Chen YG, Massague J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- Groppe JC, Shore EM, Kaplan FS. Functional modeling of the ACVR1 (R206H) mutation in FOP. Clin Orthop Relat Res. 2007;462:87–92. doi: 10.1097/BLO.0b013e318126c049. [DOI] [PubMed] [Google Scholar]
- Kaplan FS, Xu M, Glaser DL, et al. Early diagnosis of fibrodysplasia ossificans progressiva. Pediatrics. 2008;121:e1295–e1300. doi: 10.1542/peds.2007-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOP International consortium The medical management of Fibrodysplasia Ossificans Progressiva: current treatment consideration. Clin Proc Intl Clin Consort FOP. 2008;3:1–81. [Google Scholar]
- Deirmengian GK, Hebela NM, O'Connell M, Glaser DL, Shore EM, Kaplan FS. Proximal tibial osteochondromas in patients with fibrodysplasia ossificans progressiva. J Bone Joint Surg Am. 2008;90:366–374. doi: 10.2106/JBJS.G.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Thornton J. Protein fold recognition. J Comput Aided Mol Des. 1993;7:439–456. doi: 10.1007/BF02337560. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan FS, Le Merrer M, Glaser DL, et al. Fibrodysplasia ossificans progressiva. Best Pract Res Clin Rheumatol. 2008;22:191–205. doi: 10.1016/j.berh.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebela N, Shore EM, Kaplan FS. Three pairs of monozygotic twins with fibrodysplasia ossificans progressiva: the role of environment in the progression of heterotopic ossification. Clin Rev Bone Miner Metab. 2005;3:205–208. [Google Scholar]