Abstract
Down syndrome (DS) is one of the most frequent congenital birth defects, and the most common genetic cause of mental retardation. In most cases, DS results from the presence of an extra copy of chromosome 21. DS has a complex phenotype, and a major goal of DS research is to identify genotype–phenotype correlations. Cases of partial trisomy 21 and other HSA21 rearrangements associated with DS features could identify genomic regions associated with specific phenotypes. We have developed a BAC array spanning HSA21q and used array comparative genome hybridization (aCGH) to enable high-resolution mapping of pathogenic partial aneuploidies and unbalanced translocations involving HSA21. We report the identification and mapping of 30 pathogenic chromosomal aberrations of HSA21 consisting of 19 partial trisomies and 11 partial monosomies for different segments of HSA21. The breakpoints have been mapped to within ∼85 kb. The majority of the breakpoints (26 of 30) for the partial aneuploidies map within a 10-Mb region. Our data argue against a single DS critical region. We identify susceptibility regions for 25 phenotypes for DS and 27 regions for monosomy 21. However, most of these regions are still broad, and more cases are needed to narrow down the phenotypic maps to a reasonable number of candidate genomic elements per phenotype.
Keywords: Down syndrome, genotype–phenotype correlations, chromosome 21, array CGH
Introduction
Down syndrome (DS) is one of the most frequent congenital birth defects, and the most common genetic cause of mental retardation; it presents with a complex clinical spectrum of variable features affecting most organ systems. Affected individuals share certain clinical features, such as cognitive impairment, congenital heart disease and characteristic facial and physical appearance. In the vast majority of cases, DS results from the presence of an extra copy of chromosome 21.1, 2
A major goal of understanding the molecular pathology of DS is identifying genotype–phenotype correlations, that is the identification of HSA21 genes or other functional genomic elements that contribute to the specific aspects of the phenotype. There have been several general approaches to this problem: (i) mapping of partial trisomy 21 cases in human,3, 4, 5, 6, 7, 8 (ii) the construction of partial trisomy mouse models with different orthologous regions of HSA219, 10, 11 and (iii) the analysis of gene expression in cells and tissues of DS individuals or mouse models of DS, both of the transcriptome12, 13, 14, 15 and genes from the aneuploid chromosome.16, 17 Expression studies, while showing dysregulation of gene expression, have been inconclusive in identifying genes, or small HSA21 regions, for specific DS phenotypes.
The rationale for the first approach is that cases of partial trisomy 21 associated with DS features could identify genomic regions associated with specific phenotypes. Because of variable penetrance, only the presence of a particular phenotypic trait is informative for mapping. Studies of other rearrangements, including deletions and translocations, involving HSA21 may also provide information on the contribution of HSA21 genes to DS. The earliest studies hypothesized that a relatively small region of HSA21 may play a major role in DS phenotypes, and proposed the concept of a DS critical region (DSCR).4, 7, 18, 19 The DSCR was defined with a proximal boundary between markers D21S17 (35 892 kb) and D21S55 (38 012 kb), and a distal boundary at MX1 (41 720 kb).5, 8 This is a region spanning 3.8–6.5 Mb and containing ∼25–50 genes. However, analysis of further cases indicated that it was more likely that there were critical regions for particular phenotypes and not for the majority of the phenotypes.5 For example, it was previously concluded that a DS heart defect critical region maps to an ∼5.2 Mb region.6
Although the notion of a DSCR has gained some acceptance in DS research, the data supporting it remain controversial. Further, recent work on a mouse model either trisomic or monosomic for the syntenic DSCR found no evidence that the region was required to produce the characteristic facial phenotype,11 and that it was necessary, but not sufficient, for the hippocampal phenotype seen in DS.20 The limited number of partial trisomy samples, and low resolution of the mapping in earlier studies, restricted the identification of critical regions. Array comparative genome hybridization (aCGH) now allows high-resolution mapping of deletions and duplications using either oligonucleotides21 or BAC clones.22
We report here the development of a BAC array covering HSA21q to refine genotype–phenotype mapping in DS. Our study includes a considerable number of cases given the rarity of the partial aneuploidies of chromosome 21; 19 partial trisomy patients, with information on 23 phenotypic features, and 11 partial monosomy patients, with information on 27 phenotypic features were analyzed. The position of the genomic breakpoints of the partial aneuploidies was mapped to within 85 kb on average, and the results where confirmed by real-time quantitative PCR in 20 cases. We also tested five cases with a normal karyotype based on clinical findings indicative of a DS phenotype, but did not find any imbalances in chromosome 21. The minimal critical regions for certain phenotypes have been reevaluated and new boundaries have been established.
Materials and methods
Clinical samples
Patients were recruited on the basis of a chromosomal abnormality involving chromosome 21 or a phenotype with the features of DS, and the karyotypes of all 41 probands were obtained (Table 1). The phenotypes of probands were ascertained by different clinical geneticists and are reported in Tables 2 and 3. DNA samples were obtained from all cases and used for BAC array CGH.
Table 1. Summary of cases presented.
| Case | Karyotype | HAS21 Karyotype/FISH | HSA21 aCGH result | Reference |
|---|---|---|---|---|
| 1 | 47,Xinv(Y),+21 | T21 | T21 | This report |
| 2 | 47,XY,+21 | T21 | T21 | This report |
| 3 | 47,XX,+21 | T21 | T21 | This report |
| 4 | 46,t(21;21),+21 | T21 | T21 | This report |
| 5 | 46,XY,t(21;21),+21 | T21 | T21 | This report |
| 6 | 46,XY,−21, +dir dup(21)(q11.2q22.3) | PT21 | PT21 | 8, 18 and 23 |
| 7 | 46,XX, dir dup(21)(p11q22.3) | T21 | T21 | This report |
| 8 | 47,XX,+del(21)(q22.1q22.2) | PT21 | PT21 | This report |
| 9 | 47,XY,+der(21)t(3;21)(p26.1q22.12)dn | PT21 | PT21 | 24 |
| 10 | 47,XY,+der(21)t(8;21) | PT21 | PT21 | This report |
| 11 | 47,XX,+der(21),t(15;21)(q26.2;q22.1)mat | PT21 | PT21 | 8 and 25 |
| 12 | 47,XX,+der(21)(pter → q21.1::q21.3 → qter).ish der(21)(wcp21+,VIJ2yRM2029+) | PT21 | PT21 | 7 |
| 13 | 47,XY,+der(21)t(10;21)(21pter → 21q21::10q26 → 10qter)mat | PT21 | PT21 | This report |
| 14 | 46,XX, dup(21)(pter → q13::q21.3 → qter) | PT21 | PT21 | This report |
| 15 | 46,XX,−11,+der(11)t(11;21)(q24;q21) | PT21 | PT21 | This report |
| 16 | 46,XX,−10,+der(10),t(10;21)(10pter → 10q26::21q21 → 21qter)mat | PT21 | PT21 | 7 |
| 17 | 46,XX,der(21)(p11.2 → qter::q22.11 → qter) | PT21 | PT21 | This report |
| 18 | 46,XY,rec(21)dup(21q)inv(21)(p11q21) | PT21 | PT21 | This report |
| 19 | inv dup(21)(q22.3q22.1) | PT21 | PT21 | This report |
| 20 | 46,XY, der(22)(t21;22)(q22.1;q11.2) | PT21 | PT21 | This report |
| 21 | dup(21q) | PT21 | PT21 | This report |
| 22 | 46,XY,dup(21)(q22.3) | PT21 | PT21 | This report |
| 23 | 46,XX,−21,+dir dup(21)(q22.2q22.3) | PT21 | PT21 | 8 |
| 24 | 46,XX,−5,+der(5)t(5;21)(p15;q22) | PT21 | PT21 | This report |
| 25 | 46,XX,−21,+der(21)t(21;21)(p13;q22.2) | PT21 | PT21 | 8 |
| 26 | 45,XX,t(21;21)(q11;p11) | N | N | This report |
| 27 | 47,XX,+i(18)(p10) | N | N | This report |
| 28 | 46,XX | N | N | This report |
| 29 | 46,XY | N | N | This report |
| 30 | 46,XX | N | N | This report |
| 31 | 46,XX,−21,+der(15)t(15;21)(q13;q22.1) | PM21 | PM21 | 26 |
| 32 | 46,XY,−21,+del(21)(q11.1q21) | PM21 | PM21 | 27, 28 |
| 33 | 45,XY,der(2)(2pter → 2q37::21q21 → 21qter)mat,−21 | PM21 | PM21 | This report |
| 34 | 45,XX,−21/46,XX,21q- | PM21 | PM21 | This report |
| 35 | 46,XX,del(21q) | PM21 | PM21 | This report |
| 36 | 46,XX,der(21)t(3p;21q) | PM21 | PM21 | This report |
| 37 | 46,XX | PM21 | PM21 | 29 |
| 38 | 46,XX,del(21)(q21) | PM21 | PM21 | This report |
| 39 | 46,XX,del(21)(q22.3) | PM21 | PM21 | This report |
| 40 | 46,XY,der(21)t(10;21)(21pter → 21q22::10q24 → 10qter)mat | PM21 | PM21 | This report |
| 42 | 47,XXX.ish del(21q22)(AML1x1) | PM21 | PM21 | This report |
Table 2. Clinical features for trisomy and partial trisomy cases.
| Case | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | ||
| Clinical features | T21 | T21 | T21 | T21 | T21 | PT21 | T21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | PT21 | Phenotype present PT21* |
| Short stature | + | + | + | − | − | + | − | + | + | − | − | + | 5/10, 50% | |||||||||||||
| Microcephaly | + | + | − | + | − | − | 1/4, 25% | |||||||||||||||||||
| Brachycephaly | + | + | − | + | − | − | − | + | − | + | + | + | − | − | 5/12, 42% | |||||||||||
| Flat facies | + | + | − | − | − | − | + | + | 2/6, 34% | |||||||||||||||||
| Upslant palp. fissures | + | + | + | + | − | − | + | + | + | − | + | 6/9, 67% | ||||||||||||||
| Epicanthic folds | + | + | − | − | − | + | − | − | + | + | − | + | + | + | + | 7/13, 54% | ||||||||||
| Brushfield spots | − | + | − | − | − | − | + | − | 2/8, 25% | |||||||||||||||||
| Flat nasal bridge | + | + | + | − | − | − | − | + | + | + | 4/8, 50% | |||||||||||||||
| Vaulted palate | − | − | − | + | − | − | + | 2/7, 29% | ||||||||||||||||||
| Furrowed tongue | + | + | + | − | − | + | − | − | + | + | − | − | + | − | 5/12, 42% | |||||||||||
| Open mouth | + | + | + | − | − | − | − | − | − | + | + | + | + | − | − | 5/13, 39% | ||||||||||
| Malpositioned ears | − | − | − | − | − | + | + | 2/6, 34% | ||||||||||||||||||
| Small dysmorphic ears | + | + | − | + | + | − | + | + | + | + | − | 7/10, 70% | ||||||||||||||
| Cardiac anomaly | + | − | − | − | − | − | − | − | − | + | + | − | − | − | 2/12, 17% | |||||||||||
| Duodenal stenosis | − | − | − | − | 0/3, 0% | |||||||||||||||||||||
| broad short hands | + | + | + | − | − | − | + | + | + | + | + | − | 7/11, 64% | |||||||||||||
| Clinodactyly fifth finger | + | − | − | − | − | + | + | − | + | − | + | − | 5/12, 42% | |||||||||||||
| Wide-gap toes 1 and 2 | + | + | − | − | − | − | + | + | − | − | − | 4/11, 37% | ||||||||||||||
| Abnormal dermatoglyphics | − | − | − | − | + | + | − | 2/7, 29% | ||||||||||||||||||
| Palmar crease | + | − | − | − | − | + | − | + | − | 3/9, 34% | ||||||||||||||||
| Hypotonia | + | + | + | − | + | + | + | + | + | + | − | + | 8/10, 80% | |||||||||||||
| Lax ligaments | + | − | − | + | − | + | + | − | + | + | 5/9, 56% | |||||||||||||||
| IQ/MR | + | + | + | + | + | + | + | − | + | − | + | + | + | + | + | + | + | − | 13/16, 82% | |||||||
+, feature present; −, feature absent; blank, feature unknown. * The number and percentage of partial trisomy cases for a particular clinical feature where the status of the clinical feature is known.
Table 3. Clinical features for partial monosomy cases.
| Case | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical features | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 42 | Phenotype present (%)* |
| Short stature | + | + | + | − | + | 4/5 (80) | ||||||
| Short neck | + | + | − | 2/3 (67) | ||||||||
| Microcephaly | − | + | + | − | + | 3/5 (60) | ||||||
| Brachycephaly | − | 0/1 (0) | ||||||||||
| Dolichocephaly | + | − | 1/2 (50) | |||||||||
| Low hairline | + | − | 1/2 (50) | |||||||||
| Epicantic eye fold | − | − | + | + | − | 2/5 (40) | ||||||
| Hypertelorism | − | + | + | + | 3/4 (75) | |||||||
| Microphtalmia | + | − | − | 1/3 (34) | ||||||||
| Pronounced median raphe of the filtrum | + | − | 1/2 (50) | |||||||||
| Highly arched palate | + | + | − | 2/3 (67) | ||||||||
| Downslant palp.fissures | − | − | + | + | 2/4 (50) | |||||||
| Synbrachydactily | + | + | − | 2/3 (67) | ||||||||
| Brushfield spots | − | 0/1 (0) | ||||||||||
| Flat nasal bridge | − | − | 0/2 (0) | |||||||||
| Broad nasal bridge | + | + | − | − | 2/4 (50) | |||||||
| Broad mouth | + | + | − | + | 3/4 (75) | |||||||
| Large ears | + | + | − | + | 3/4 (75) | |||||||
| Large nose | + | + | + | − | 3/4 (75) | |||||||
| Cardiac anomaly | − | − | − | + | 1/4 (25) | |||||||
| Clinodactily of fifth finger | − | + | − | + | 2/4 (50) | |||||||
| Palmar crease | − | + | − | 1/3 (34) | ||||||||
| Hypotonia | + | − | − | 1/3 (34) | ||||||||
| Hypertonia | + | + | − | − | 2/4 (50) | |||||||
| Seizure | + | + | − | 2/3 (67) | ||||||||
| Lax. ligaments | − | − | 0/2 (0) | |||||||||
| IQ or MR | + | + | + | + | + | + | + | 7/7 (100) | ||||
+, feature present; −, feature absent; blank, feature unknown. *The number and percentage of partial monosomy cases for a particular clinical feature where the status of the clinical feature is known.
Cases 1–25 are patients with DS features according to the criteria of Jackson et al23 (Table 2). Cases 1–3 are complete trisomy 21 and were included as controls for array CGH. Cases 4, 5 and 26 involve translocations of HSA21, and were examined to see whether the translocations are balanced. Cases 6, 7 and 8–25 are partial trisomies for chromosome 21; the origins of partial trisomy are de novo direct duplication, de novo translocation or missegregation of a parental balanced translocation. We include clinical data of 18 cases (data for cases 1, 2, 4, 7, 21, 22 and 24 are unavailable). For cases with partial trisomy, features considered to be common in DS were evaluated wherever possible (Table 2).
Cases 31–42 are patients with partial monosomy 21 (Tables 1 and 3). Despite the phenotypic variability in partial monosomy 21, there are several common features including craniofacial, skeletal and cardiac defects, genital malformations and severe mental retardation.24, 25, 26, 27, 28, 29, 30, 31, 32, 33 All ascertained partial monosomy cases presented with mild-to-severe mental retardation.
Cases 26–30 were included because of a DS-like phenotype. Case 27 presented with a DS-like phenotype, including microcephaly, upslanted palpebral fissures, thin philtrum, camptodactyly and moderate mental retardation. Karyotype analysis indicates a normal HSA21, but tetrasomy 18p. Case 28 is a patient with relative microcephaly, failure to thrive, developmental and speech delays, midline cleft palate, pharyngeal dysfunction, gastroesophageal reflux, strabismus and farsightedness. Case 29 presented with microcephaly, brachycephaly, low posterior hairline, scarce and flared eyebrows, upslanted palpebral fissures, malar hypoplasia, small nares, small ears, thin philtrum, down-turned corners of the mouse, small mouth, tongue with midline groove, crowded teeth, sacral dimple, fifth finger clinodactyly and developmental delay. The result of karyotype analysis is 46, XX. Case 30 was referred for mental retardation, heart malformation and dysmorphism at the age of 6 years. At the first evaluation, he presented a mild-to-moderate mental retardation with speech delay, hyperactivity and dysmorphic features, including midface hypoplasia, small ears, intermittent convergent strabismus, short upturned nose with anteverted nares. Small hands were also present with normal skin creases and clinodactyly of the fifth toes. A large ventricular septal defect was discovered at the age of 2 months. Cytogenetic analysis reported normal karyotype 46,XY, and no subtelomeric abnormality was detected by MLPA.
Preparation of an HSA21q BAC array
A total of 409 BACs were selected to cover the long arm of human chromosome 21. The BACs have a mean length of 157 kb and a mean overlap of 85 kb giving an ∼twofold tiling path (Supplementary Figure 1). There are seven gaps (range: 9449–297 923 bp, mean 101 653 bp) because of the lack of spanning BAC clones in the libraries used. Sixty-four BACs from other chromosomes were used as normalization controls. BACs where obtained from libraries RPCI-11 (CHORI, www.chori.org/bacpac) and CTD (California Institute of Technology).34 The majority of BACs were obtained as DNA from CHORI. For BACs obtained as cultures, DNA was prepared using the Montage BAC96 kit from (Millipore) on a Beckman 2000 robot. All BAC DNAs were amplified by DOP PCR,22 and diluted to 200 ng μl−1 in Nexterion Spot I (Schott) spotting buffer. BACs were printed in duplicate onto Nexterion AL slides (Schott) using an SDDC-2 ESI robot (BioRad). After printing, slides were incubated for 15 min in a humid chamber, 1 h at 120 °C, and then stored in a desiccator.
Array hybridization and washing
In all, 600 ng of DNA was labeled with Cy3 or Cy5 (Amersham) using the BioPrime labeling kit (Invitrogen) following the manufacturer's instructions. Array hybridization and washing was performed according to Fiegler et al22 except that probe denaturation and prehybridization were at 70 °C instead of 72 °C.
Array analysis
Raw data were obtained from hybridized arrays using a GSI Lumonics ScanArray4000 scanner and ImaGene software (BioDiscovery). Analysis was performed in two stages. First, duplicate BAC spot values were averaged, and then all values were normalized to the mean of all control (non-HSA21) BACs. Second, samples were analyzed using CGH-explorer35 to determine ploidy and to identify breakpoints. Calculations were carried out in Excel (Microsoft Corporation), and statistics and graphing in R (www.R-project.org).
Genotype–phenotype mapping
To map phenotypes along HSA21, we used a scoring system for each BAC, as follows. A binary scoring system was used because, although DS phenotypes are quantitative, we only have data on presence or absence of the phenotype. First, we considered only cases with the presence of a particular phenotype (Figures 3 and 4). Each BAC was given a value of 1, if it was trisomic in the presence of the phenotype, or a value −1, if euploid in the presence of the phenotype. For each phenotype, a score for each BAC was then calculated by summing the values for those cases where the phenotype is present. Similarly, for monosomy samples, each BAC was given a value of 1 for monosomy or −1 for euploid, and each BAC scored for phenotype by summing the positive cases. Second, to try to include penetrance in the analysis, we adjusted the scores to take into consideration trisomic cases, which do not have the phenotype (Supplementary Figure S2). This was carried out by giving a value of −1 to BACs trisomic in cases without the phenotype and summing across all cases (both presence and absence of the phenotype).
Quantitative PCR
SYBR green assays were designed using the program PrimerExpress (Applied Biosystems) with default parameters in every case. Repetitive sequences were masked using REPEATMASKER (www.repeatmasker.org), and amplicon sequences were checked by BLAT36 against the human genome to ensure that they were specific for the region under study. All reactions used 300 n of each primer, 10 ng of genomic DNA and PowerSybrGreen MasterMix (AppliedBiosystems). PCRs were set up using a Biomek 2000 robot (Beckman), in a 10-μl volume in 384-well plates with two replicates per sample. Reactions were run in an ABI 7900 Sequence Detection System (Applied Biosystems) with the following conditions: 50 °C for 2 mins, 95 °C for 10 mins and 50 cycles of 95 °C 15 s per 60 °C 1 min. For data analysis, Ct values were obtained using SDS 2.0 (Applied Biosystems), input DNA quantities were normalized to four assays from chromosome 10 and relative DNA copy number obtained by pairwise comparisons of test and three control DNAs. Calculations were carried out in Excel (Microsoft Corporation) and graphing in R (www.R-project.org).
Results and discussion
We present 41 cases in this study, including 30 with partial aneuploidy for chromosome 21 (Table 1): 19 cases of partial trisomy 21 (12 new and seven previously reported cases)7, 8, 18, 37, 38, 39 and 11 cases of partial monosomy 21 (eight new and three previously reported cases).24, 40, 41, 42 We also studied five cases presenting a DS-like phenotype with a normal karyotype. The remaining six trisomy 21 cases consisted of two t(21;21) and one duplication involving HSA21 tested to identify possible partial trisomy and three free trisomy 21 cases used as controls. The clinical features of the 41 cases were collected from the original medical records given by the referring physician. The available data and clinical evaluations are summarized in Tables 2 and 3.
All 41 cases were analyzed by aCGH on the HSA21q BAC array (Figures 1 and 2). The results by aCGH were concordant with cytogenetic observation in all cases. Validation by real-time quantitative PCR was carried out for 20/30 (64%) of cases with partial aneuploidy, and confirmed the aCGH results (Figure 1c).
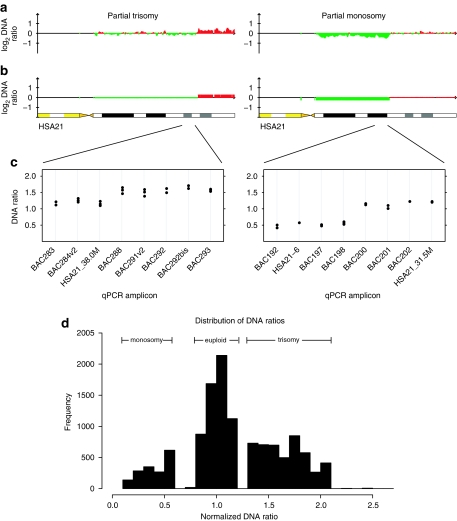
Figure 1.
Examples of HSA21 array CGH results. (a) Plots of DNA ratio for two cases. X-axis, position along HSA21; Y-axis, normalized log2 ratio of case/control signal. (b) Data from a segment using CGH-explorer.35 The examples show cases of partial trisomy (left panels) and partial monosomy (right panels). (c) Confirmation of results and refinement of breakpoints by quantitative PCR. X-axis, quantitative PCR amplicon (see Supplementary Table 1 for locations of assays), Y-axis and normalized ratio of case/control signal. Each assay is represented by three points, the case compared with three different control DNAs. (d) Histogram of the distribution of all normalized case/control DNA ratios, total ≈16 000 data points.
Figure 2.
aCGH results. X-axis represents position along the HSA21q in Mb; Y-axis represents the cases. Shading represents ploidy along the chromosome as defined in the key. Two array clone gaps are visible in each sample.
We investigated four cases (27–30), which presented with the aspects of a DS phenotype, two of which had karyotypes with abnormalities involving HSA21. Case 27 was investigated because of the presence of several features of DS, and a tetrasomy 18p karyotype, which might have masked a cryptic trisomy 21. However, aCGH revealed a normal HSA21 content. Cases 28–30 had some features of DS leading us to suspect a possible chromosome 21 cryptic duplication. Again, aCGH analyses revealed a normal HSA21 content for all three cases. Case 26 was included as normal control with a balanced t(21;21) and normal phenotype.
Despite the phenotypic variability between cases of full and partial monosomy 21 reported in the literature, features are frequently described include intrauterine prenatal and postnatal growth retardation, down-slanted palpebral fissures, low set ears, hypertonia, heart defect and mental retardation.6 We report the clinical findings of 7/12 cases for partial monosomy 21. All the cases reported (7/7) were described with mental retardation. Short stature is present in 4/5 cases of partial monosomy 21. Large ears, large nose, broad mouth and hypertelorism are frequently associated with partial monosomy 21, each with 3/4 cases presenting the phenotype when assessed.
Case 39 was reported with subtle dysmorphic anomalies including marfanoid habitus. The thin marfanoid build was previously associated with distal monosomy 21q.4, 24
The breakpoints of the different partial aneuploidies appear to be nonrandomly distributed along the length of 21p. If one divides 21q in the bins of 5 Mb, the segments between 30–35 Mbs and 35–40 Mbs contain 10 and 16 breakpoints (Figure 2), respectively; in contrast, all other such segments contain 0–6 breakpoints each. This distribution of breakpoints result in a P value=0.0064 (Fisher's exact test). This could be the result of an ascertainment bias because the cases have been collected based on phenotypic consequences; alternatively, this could reflect a differential propensity of DNA sequences for breakage.
Partial monosomy 21
We present eight new cases of partial monosomy (cases 33–36, 38–40 and 42) and a refinement of the mapping for three previously reported cases (31,40 3224, 41 and 3742). Cases 31 and 32, presenting a deletion of the proximal region of chromosome 21, were described with a severe phenotype including severe mental retardation, craniofacial abnormalities (broad forehead, downward-slanting palpebral fissures and low set, large ears). The 11 partial monosomies ranged in size from 1.48 Mb (case 42) to 21.06 Mb (case 31). All the partial monosomies were unique as none of the patients seems to share a common breakpoint (Figure 4). The smallest deletion (case 42) was estimated by aCGH to be only 1.48 Mb (Figure 4), and contains eight genes including DSCR1 and RUNX1. Case 42 has a relatively severe phenotype including mental retardation, microcephaly, short stature and cardiac anomaly.
The largest deletions mapped on chromosome 21 are 18.20 Mb (case 31) and 17.51 Mb (case 32) between the centromere and 21q22.11. Both of these cases have a severe phenotype including severe mental retardation, craniofacial abnormalities (broad forehead, downward-slanting palpebral fissures and low set, large ears). Cases 37 and 38 with distal deletions ranging from 11 Mb (case 35) to 5.63 Mb (case 37) have a relatively milder phenotype including moderate mental retardation and absence of craniofacial anomalies. Case 39 was reported with subtle dysmorphic anomalies including marfanoid habitus. The thin marfanoid build was previously associated with distal monosomy 21q.26, 43
The partial monosomy cases described here indicate how deletion of three broad regions of HSA21 contribute to the phenotype of monosomy. The first region, from the centromere to ∼31.2 Mb produces a severe phenotype. This covers the gene-poor region of HSA21, but contains ∼50 genes. In the second region from 31.2–36 Mb, there are no cases with a deletion spanning this region, and only one case (42) with a partial deletion of this region (case 42), and this case has a severe phenotype. This indicates that this region, which has a high gene density (∼80 genes), contains a combination of genes that may not be tolerated in a monosomic state. The third region, from ∼36–37.5 Mb to the telomere, contains a large number of genes ∼130, but its monosomy results in a milder phenotype.
These data agree with previously published cases of partial trisomy 21.24, 27, 28, 29 For example, Chettouh et al24 presented six cases, four of whom had large proximal deletions and a severe phenotype, similar to cases 31 and 32 in our study. Also, Ehling et al27 presented two cases with distal deletions and a mild phenotype, consistent with cases presented here.
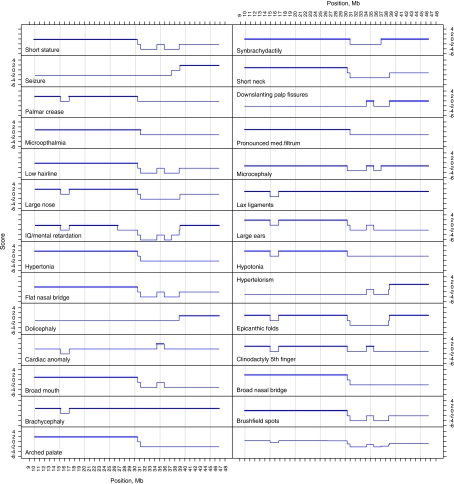
Given the rarity of partial monosomies, it is difficult to draw firm conclusions. However, for monosomy phenotypes, it seems that there is no region which would correspond to a ‘critical region' (Figure 3). The cases presented here and in other studies24, 27, 28, 29 show that many of the individual phenotypes can be present when different regions of HSA21 are deleted.
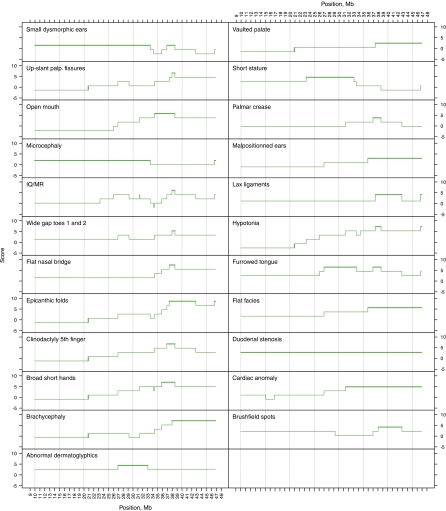
Figure 3.
Genotype–phenotype mapping in partial trisomy 21. Each graph represents one aspect of the phenotype (Table 2). The X-axis represents the position along HSA21q in Mb; Y-axis is the phenotype score for each BAC, with the maximal region shown in bold.
Partial trisomy 21 and critical regions
The 19 partial trisomies reported here range in size from 5.98 Mb (case 23) to 28.56 Mb (case 6), and each case is unique as none of the patients share a common breakpoint (Figure 2). The clinical features of the cases included in this study were collected from the original medical records provided by the referring physicians. We evaluated the most frequent clinical feature described for the 19 cases of partial trisomies, and the available data and clinical evaluations are summarized in Table 2.
DS affects multiple systems and produces both functional and structural defects. T21 is frequently associated with mental retardation, congenital heart defects (mainly atrioventricular septal defect), abnormalities of the gastrointestinal tract, abnormalities of neuromuscular tone, characteristic facial and physical features, a high incidence of seizures, modified audiovestibular and visual functions and the early onset of Alzheimer's disease (AD). To date, almost every aspect of the phenotype of DS is subject to high degree of variability even in cases of full trisomy 21. Only two of these features are observed in all DS patients: mental retardation and neuropathological modifications similar to those observed in the brains of AD patients (in DS patients over the age of 35 years).
Typically, DS patients exhibit a progressive decline in IQ beginning in the first year of life. By adulthood, IQ is usually in moderate-to severe retardation ranged (IQ=25–55) with an upper limit on mental age of ∼7–8 years, although a few individuals have IQ in the lower normal range (70–80 years).44 We report 13/16 (82%) cases with mental retardation in patient with partial trisomy 21, a lower rate compared to the value of 100% described in DS patients with full trisomy 21. The minimum region defined in our study maps is between 37.94 and 38.64 and contains KCNJ6, DSCR4 and KCNJ15. However, cases 9, 10, 11 and 13 define a second region from the centromere to 26.96 contributing to MR.
Hypotonia, which is frequently observed in neonates, is difficult to associate with a well characterized developmental anomaly,45 but is reported as the most frequent sign of DS.1 In our study, hypotonia was present in 80% of the partial trisomy 21 cases, and enables the mapping of hypotonia region to two small genomic segments 37.4–38.4 and 46.5-qter.
The mapping of the triplicated genomic segment of chromosome 21 that harbors the functional elements contributing to congenital heart defect (CHD)46, 47 is of importance to understand the pathogenesis of these anomalies. CHD is present in only 2/12 (17%) compared to the overall risk of CHD of 40% reported in DS patients. Earlier studies of rare individuals with CHD and partial duplications of chromosome 21 established a candidate region from D21S3 to PFKL,6 which agrees with the two cases presented here (Figure 3). However, the sample in our study maps the CHD region in a large genomic segment between 31.5 Mb and qter.
Earlier data show that not all the DS patient display microcephaly and the main feature observed is brachycephaly. Our study on partial trisomy 21 shows that 25 and 42% of partial trisomy 21 were reported with microcephaly and brachycephaly, respectively. Major craniofacial abnormalities well described in DS were reported with variable rate in our study; for example, upslanting palpebral fissure is present in 67% of the cases, flat facies in 34% and brushfield spots in 25% of the cases. Figure 3 shows the extent of the phenotypic mapping positions of these features.
In summary, partial trisomy patients display particular phenotypes at a lower frequency than trisomy 21 patients (Table 2).
Genotype–phenotype correlations
The goal for mapping phenotypes to specific regions of HSA21 is to identify which genes (or small regions) contribute to DS phenotypic features, and thus to understand DS pathogenesis.48 Phenotype candidate regions are defined as the minimum region of overlap between cases positive for the phenotype. To map these regions, we calculated a score for each BAC along HSA21 (see Materials and methods for details). The scores are plotted in Figures 3 and 4, highlighting the highest scoring regions for each phenotype. Clearly, the majority of the phenotypes map to distal HSA21; averaging the phenotype score indicates a region ∼37–44 Mb, which is involved in most DS phenotypes. This is not surprising, as this is the most gene-rich region of chromosome 21.49
Figure 4.
Genotype–phenotype mapping in partial monosomy 21. Each graph represents one aspect of the phenotype (Table 3). The X-axis represents the position along HSA21q in Mb; Y-axis is the phenotype score for each BAC, with the maximal region shown in bold.
Confidence in these scores can be ascertained by assessing how many total cases express the phenotype compared to those with no phonotype (ie, penetrance; see the last column of Table 3). Adjusting the scoring system described above for each BAC was done to take this type of data into consideration (see Materials and methods; Supplementary Figure S2). For example, cases 16 and 18 have cardiac anomaly, and the line (Figure 3) represents the region of overlap between the trisomic regions for these two cases (Figure 2). However, in addition to these data, we have data from many more cases (8, 9, 10, 11, 12, 20, 23 and 25) that do not have cardiac anomaly, but are also trisomic for at least a part of this region. For many phenotypes, this analysis may reduce the size of the candidate regions, but it has the problem that it does not take into account the complicating factor of reduced penetrance (Supplementary Figure S2).
Cases 9, 10, 11, 12 and 13 are interesting as they are trisomic for proximal HSA21 and do not include the ‘DSCR' (Table 1). These cases, and three other similar cases reported previously,50 exclude the possibility of there being a single DSCR, in the sense of a single region being responsible for all aspects of the phenotype.
Although the most likely region for many phenotypes maps in 34–41 Mb, other regions are important for microcephaly, abnormal dermatoglyphics, short stature and furrowed tongue. Gene density correlates with DS (average) phenotype (Spearman rho 0.58, P<0.0001), and this indicates that many genes along HSA21 make a contribution to the overall DS phenotype.
For a true DSCR, there must be no patients who have the major features of DS but do not have this region. Rather this is a susceptibility region (SR) modified by other loci on HSA21 and elsewhere in the genome. The DSCR may thus be described as a phenotype SR. These SRs make sense against a background of expression variation.51, 52
Mouse partial trisomies that are syntenic to trisomy 21 in humans provide important information regarding the genome mapping of phenotypic characteristics. For example, trisomy syntenic for the ‘DSCR' named Ts1Rhr11 did not display learning and memory abnormalities or facial dysmorphism, and thus did not provide evidence for the ‘DSCR'. However, genetic differences between strains and the problem of comparing phenotypes between mouse and human are an issue for DS mouse models in general. As in many other biological investigations, the positive result is significant, whereas the negative is uninformative.
Many more additional studies are needed to reduce the candidate regions for certain phenotypes. Given the rarity of these partial aneuploidies for chromosome 21, collection of cases from several centers need to be centralized and analyzed with a common diagnostic platform. In addition, a careful and structured evaluation of the phenotypic characteristics will result in meaningful comparisons and conclusions. The use of oligonucleotide platforms for array comparative genomic hybridization for diagnostic purposes may reveal a considerable number of additional cases that could otherwise be undiagnosed. There is a need for a web-based reporting and collection of both phenotypic and genotypic characterization.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (RL and SEA), the NCCR Frontiers in Genetics (SEA), the European Union (AnEUploidy to SEA), Child Care foundation (SEA), the Fondation Jérôme Lejeune (AP).
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Epstein CJ.Down syndrome (trisomy 21)in Scriver CR, Beaudet AL, Sly WS, Valle D (eds):New York: McGraw-Hill; 20011223–1256. [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Bradley C, Disteche CM. Down syndrome: molecular mapping of the congenital heart disease and duodenal stenosis. Am J Hum Genet. 1992;50:294. [PMC free article] [PubMed] [Google Scholar]
- Sinet PM, Theophile D, Rahmani Z, et al. Mapping of the Down syndrome phenotype on chromosome 21 at the molecular level. Biomed Pharmacother. 1994;48:247. doi: 10.1016/0753-3322(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Schipper R, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow GM, Chen XN, Shi ZY, et al. Down syndrome congenital heart disease: a narrowed region and a candidate gene. Genet Med. 2001;3:91. doi: 10.1097/00125817-200103000-00002. [DOI] [PubMed] [Google Scholar]
- McCormick MK, Schinzel A, Petersen MB, et al. Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics. 1989;5:325. doi: 10.1016/0888-7543(89)90065-7. [DOI] [PubMed] [Google Scholar]
- Delabar JM, Theophile D, Rahmani Z, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, et al. A mouse model for Down syndrome exhibits learning and behavioural deficits. Nat Genet. 1995;11:177. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA. 1998;95:6256. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific down syndrome phenotypes. Science S. 2004;306:687. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, et al. The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res. 2000;10:2006. doi: 10.1101/gr.10.12.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum Mol Genet. 2002;11:3249. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- Mao R, Zielke CL, Ronald Zielke H, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing down syndrome brain. Genomics. 2003;81:457. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Saran NG, Pletcher MT, Natale JE, Cheng Y, Reeves RH. Global disruption of the cerebellar transcriptome in a down syndrome mouse model. Hum Mol Genet. 2003;12:2013. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis ES. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 2004;14:1268. doi: 10.1101/gr.2090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlem P, Sultan M, Herwig R, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res S. 2004;14:1258. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani Z, Blouin JL, Creau-Goldberg N, et al. Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc Natl Acad Sci USA. 1989;86:5958. doi: 10.1073/pnas.86.15.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg JR, Kawashima H, Pulst SM, et al. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am J Hum Genet. 1990;47:236. [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Roper RJ, Sengstaken CL, et al. Trisomy for the Down syndrome “critical region” is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet. 2007;16:774–782. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Jackson JF, North ERr, Thomas JG. Clinical diagnosis of Down's syndrome. Clin Genet S. 1976;9:483. doi: 10.1111/j.1399-0004.1976.tb01601.x. [DOI] [PubMed] [Google Scholar]
- Chettouh Z, Croquette MF, Delobel B, et al. Molecular mapping of 21 features associated with partial monosomy 21: involvement of the APP-SOD1 region. Am J Hum Genet. 1995;57:62. [PMC free article] [PubMed] [Google Scholar]
- Barnicoat AJ, Bonneau JL, Boyd E, et al. Down syndrome with partial duplication and del (21) syndrome: study protocol and call for collaboration. Study I: clinical assessment. Clin Genet. 1996;49:20. doi: 10.1111/j.1399-0004.1996.tb04319.x. [DOI] [PubMed] [Google Scholar]
- Bartsch O, Petersen MB, Stuhlmann I, et al. “Compensatory” uniparental disomy of chromosome 21 in two cases. J Med Genet. 1994;31:534. doi: 10.1136/jmg.31.7.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling D, Kennerknecht I, Junge A, et al. Mild phenotype in two unrelated patients with a partial deletion of 21q22.2-q22.3 defined by FISH and molecular studies. Am J Med Genet A. 2004;131:265. doi: 10.1002/ajmg.a.30361. [DOI] [PubMed] [Google Scholar]
- Huret JL, Leonard C, Chery M, et al. Monosomy 21q: two cases of del(21q) and review of the literature. Clin Genet. 1995;48:140. doi: 10.1111/j.1399-0004.1995.tb04074.x. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Kalousek DK, Anneren G, et al. Deletion of chromosome 21 and normal intelligence: molecular definition of the lesion. Hum Genet. 1991;87:112. doi: 10.1007/BF00204163. [DOI] [PubMed] [Google Scholar]
- Nielsen F, Tranebjaerg L. A case of partial monosomy 21q22.2 associated with Rieger's syndrome. J Med Genet. 1984;21:218. doi: 10.1136/jmg.21.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos DS, Cowan JM, Elias ER, Cole C. Physical findings in 21q22 deletion suggest critical region for 21q- phenotype in q22. Am J Med Genet. 1995;59:161. doi: 10.1002/ajmg.1320590209. [DOI] [PubMed] [Google Scholar]
- Valero R, Marfany G, Gil-Benso R, et al. Molecular characterisation of partial chromosome 21 aneuploidies by fluorescent PCR. J Med Genet. 1999;36:694. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Ogasawara N, Gotoh A, Komiya H, Nakai H, Kuroki Y. A case of 21q – syndrome with normal SOD-1 activity. Hum Genet. 1979;48:321. doi: 10.1007/BF00272832. [DOI] [PubMed] [Google Scholar]
- Kim UJ, Birren BW, Slepak T, et al. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Lingjaerde OC, Baumbusch LO, Liestol K, Glad IK, Borresen-Dale AL. CGH-Explorer: a program for analysis of array-CGH data. Bioinformatics. 2005;21:821. doi: 10.1093/bioinformatics/bti113. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT – The BLAST-like alignment tool. Genome Res. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin JL, Aurias A, Creau-Goldberg N, et al. Cytogenetic and molecular analysis of a de novo tandem duplication of chromosome 21. Hum Genet. 1991;88:167. doi: 10.1007/BF00206066. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Mizuno S, Ohara K, et al. Two cases of partial trisomy 21 (pter-q22.1) without the major features of Down syndrome. Am J Med Genet A S. 2006;140:227. doi: 10.1002/ajmg.a.31073. [DOI] [PubMed] [Google Scholar]
- Raoul O, Carpentier S, Dutrillaux B, Mallet R, Lejeune J. Partial trisomy of chromosome 21 by maternal translocation t(15;21) (q26.2; q21) Ann Genet. 1976;19:187. [PubMed] [Google Scholar]
- Rethore MO, Dutrillaux B. Translocation 46,XX, t(15; 21) (q13; q22,1) in the mother of 2 children with partial trisomy 15 and monosomy 21. Ann Genet. 1973;16:271. [PubMed] [Google Scholar]
- Orti R, Megarbane A, Maunoury C, Van Broeckhoven C, Sinet PM, Delabar JM. High-resolution physical mapping of a 6.7-Mb YAC contig spanning a region critical for the monosomy 21 phenotype in 21q21.3-q22.1. Genomics. 1997;43:25. doi: 10.1006/geno.1997.4765. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M, Vestermark S. Karyotype 45,XX,-21/46,XX,21q-in an infant with symptoms of G-deletion syndrome I. J Med Genet. 1974;11:389. doi: 10.1136/jmg.11.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Moon J, Edgin J, Stedron J, Nadel L. The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child Dev. 2003;74:75. doi: 10.1111/1467-8624.00522. [DOI] [PubMed] [Google Scholar]
- Delabar JM, Aflalo-Rattenbac R, Creau N. Developmental defects in trisomy 21 and mouse models. ScientificWorldJournal. 2006;6:1945. doi: 10.1100/tsw.2006.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubman TR, Shields MD, Craig BG, Mulholland HC, Nevin NC. Congenital heart disease in Down's syndrome: two-year prospective early screening study. BMJ. 1991;302:1425. doi: 10.1136/bmj.302.6790.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torfs CP, Christianson RE. Anomalies in Down syndrome individuals in a large population-based registry. Am J Med Genet. 1998;77:431. [PubMed] [Google Scholar]
- Epstein CJ, Korenberg JR, Anneren G, et al. Protocols to establish genotype-phenotype correlations in Down syndrome. Am j Hum Genet. 1991;49:207. [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Schipper R, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini P, Deutsch S, Lyle R, et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet. 2007;81:252. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait Yahya-Graison E, Aubert J, Dauphinot L, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet. 2007;81:475. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.