Abstract
Adolescent idiopathic scoliosis (AIS) is widely recognized as a complex disorder with a strong genetic predisposition. In previous studies, a number of extracellular matrixes (ECMs) related genes have been duplicated as candidate genes for AIS. Matrilin-1 plays an important role in the organization of the ECM, and matrilin-1 gene (MATN1) mutant mice showed similar phenotypes to scoliosis. We hypothesized that MATN1 was a candidate predisposition gene for AIS. A gene-based association study was conducted using seven tagging SNPs identified from the HapMap data. For initial screening, the seven tagSNPs were genotyped in 197 cases and 172 controls. Next, we validated any significant association in an additional sample of 222 cases and 288 controls. In addition, another 290 controls were genotyped to confirm the results. We found that allele G of rs1149048 was a significant predisposition allele of AIS (P=0.0007, odds ratio (OR)=1.35 within 95% confidence interval (CI)=1.14–1.61), and individuals with genotype GG had a higher risk for AIS compared with AA+AG (P=0.0001, OR=1.61 within 95% CI=1.25–2.08). Polymorphism of rs1149048 was also associated with curve severity in AIS patients. Also, a significantly higher maximum Cobb angle was found in patients with GG genotype (P=0.002). We concluded that the tagSNP rs1149048 polymorphism in the MATN1 promoter region was associated with both susceptibility and disease progression in AIS.
Keywords: scoliosis, tagging SNP, polymorphism, MATN1
Introduction
Adolescent idiopathic scoliosis (AIS) is the most common spine deformity during puberty, with a prevalence of 2–4% in adolescent population,1 and poses a significant health burden to children. The etiology of AIS has not been fully understood. Many hypotheses have been proposed, including genetic factors, hormonal disturbance, effects of connective tissue and abnormal growth.2 It is widely recognized that hereditary or genetic factors play an important role in the development of AIS. Clinical observations as well as population studies have documented scoliosis within families, with the prevalence higher among relatives than within the general population.2, 3
The mode of inheritance is still unclear, but evidence for autosomal dominant4 X-linked inheritance5 and multifactorial mode of inheritance has been described. Genome-wide linkage studies have identified several loci predisposing to AIS. Chromosomal regions on 6q, 10q and 18q,6 17p11.2,7 19p13.3,8, 9 8q11,10 Xq23–26.1,5 9q31.2–q34.2 and 17q25.3–qtel11 have been reported. A genomic screen of a large sample of families by Miller et al12 investigated five primary (chromosomes 6p, 6q, 9, 16 and 17) and eight secondary (chromosomes 1, 3, 5, 7, 8, 11, 12 and 19) areas of significance. Also, the locus on chromosome 19p13 described earlier9 has been independently confirmed.8 These studies indicate that AIS may be a complex genetic disorder that is the result of one or more genetic loci with complex genetic–environment interactions.
Genetic association study, which is more powerful to detect genetic contributions to complex disease than linkage studies,13 has recently been used to study genetic predisposition in AIS. Polymorphisms associated with AIS have been described in SNTG1 on 8q11.22,10 ESR1 on 6q25.1,14 MATN1 on 1p35,15 CHD7 on 8q12.116 and MTNR1B on 11q21–q22.17 Genetic association study using a case-only sample may reveal disease-modifying genes that influence disease progression.13 Using a case-only study, Inoue et al18 reported an association between the XbaI polymorphism of ESR1 gene and curve severity of AIS. The case-only study design indicated that XbaI polymorphism was a disease modifier gene in AIS.
Extracellular matrixes (ECMs) are principal elements in the supporting structures of the spinal column and have been the focus of many studies dealing with the pathophysiology of idiopathic scoliosis.2 As scoliosis is a phenotypic characteristic of many ECM disorders, such as Marfan syndrome and Ehlers–Danlos syndrome, the hypothesis that a defect within the ECM is the causative factor of idiopathic scoliosis is plausible. Preliminary studies have focused on the genes responsible for the structural components of the ECM system, including COL1A1,19 COL1A2,19, 20 COL2A1,19 FBN1,20 elastin20 and aggrecan genes;21, 22 however, all the genes were excluded as potential causative factors for idiopathic scoliosis within the study populations.
Matrilin-1, a non-collagenous protein, is secreted primarily by chondrocytes and has a role in the assembly of cartilage ECM.23 It has been confirmed that matrilin-1 has an important function in the organization of chondrocyte into distinct zones of growth plate.24 Disturbance of the chondrocyte zonal distribution could lead to musculoskeletal disorders,25 such as scoliosis. Identification of a mutation in matrilin-3 in patients suffering from multiple epiphyseal dysplasia26 demonstrated the importance of matrilins in cartilage development. By reviewing mouse phenotypes exhibiting scoliosis, kyphosis or tail kinks, matrilin-1 gene was proposed as a candidate gene related to human scoliosis on the basis of synteny conservation.27, 28 Moreover, a recent study15 showed that an intragenic microsatellite (STRP) polymorphism in the 3′ untranslated region (3′-UTR) of MATN1 is associated with AIS in a TDT study of 81 trios. These data provided strong evidence that MATN1 could be a good candidate gene for AIS.
In this study, we aim to examine the genetic association between MATN1 and AIS. To address this issue, we applied the recently recommended strategy of gene-based association analysis.13, 29 We conducted a case–control association analysis in a Chinese population by selecting tagging SNPs (tSNPs) from the HapMap database.30
Materials and methods
Subjects
This study population consisted of 419 patients with AIS (89.8% female, average age 16.1±0.93 years, range 12–19 years) and 750 healthy controls (88.3% female, average age 15.8±0.10 years, range 8–24 years); all were Chinese. All patients were seen as at the Spine Surgery of Drum Tower Hospital of Nanjing University Medical School between 2005 and 2007. Of these patients, 103 received operation and 152 received brace treatment, whereas 164 patients received no treatment other than regular clinical observation.
The effect of gene polymorphisms on the curve severity of AIS was also analyzed. As bracing could change the natural history of AIS, patients with bracing treatment were excluded from analysis. So a subgroup of AIS patients (N=267) who received continued observation or surgery were included. All the patients had been followed more than 1 year until the age of growth maturation (≥16 years old), and the maximum Cobb angle was recorded as either the last Cobb angle at 16 years of age for non-operated cases or the latest Cobb angle before surgery.
All subjects of the study met the following inclusion criteria: no evidence of bone diseases, metabolic diseases or growth disturbances; no evidence of systemic illness or other condition known to affect bone metabolism; and no history of recent steroid intake. Informed consent to DNA analysis was obtained from all subjects and/or their parents. The Hospital Ethical Committee approved the study.
Characterizing linkage disequilibrium and tagging SNP selection
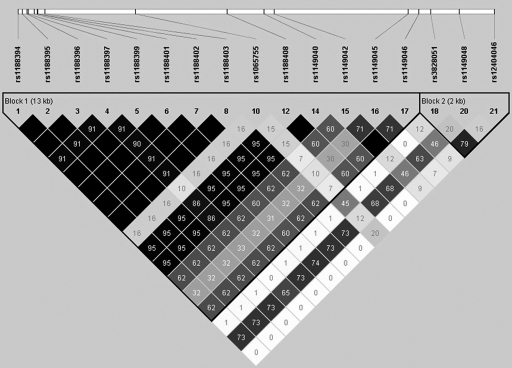
SNPs were retrieved from HapMap database for Han population sample (release no. 21a/phase II, population: CHB, MAFs>0.1). Seventeen SNPs covering 17.89 kb including MATN1 (spanning from 5 kb upstream to 2 kb downstream of the gene) were identified (Figure 1). Seven tSNPs (SNP1: rs1188402, SNP2: rs1065755, SNP3: rs1149045, SNP4: rs1149046, SNP5: rs3828051, SNP6: rs1149048 and SNP7: rs124040046) were selected, R2≥0.831 for the following association analysis.
Figure 1.
Linkage disequilibrium (LD) structure of region around MATN1. Two LD blocks were reconstructed by HapMap Phase II Han Chinese population data containing 17 SNPs spanning from 5 kb upstream to 2 kb downstream of MATN1. Pair-wise LD value of R2 is represented by the degree of shading.
SNP genotyping
Genomic DNA was prepared from peripheral blood leukocytes by using the chelex-100 method.32 Genotypings were carried out by PCR-RFLP analysis. Primers were designed with Primer 3 software.33 The primers are listed in Table 1. The reaction mixture was subjected to denaturation at 96°C for 2 min, followed by 30 cycles at 94°C for 1 min, 60–64°C for 1 min, 72°C for 1 min, then by a final extension at 72°C for 15 min. After digestion with an appropriate restriction enzyme, PCR products were electrophoresed on a 2% agarose gel and visualized by ethidium bromide staining. To validate the genotyping results, duplicated genotyping was performed in 10% of the samples to confirm the results.
Table 1. Primers and restriction enzymes for RFLP analysis in this study.
| dbSNP ID | Location | Nucleotide changea | Primers | Enzyme |
|---|---|---|---|---|
| rs1188402 | 3′-flanking | +11978 A>G | F: 5′-GGACAGAGAGAAAGGGTTTGG-3′ | AluI |
| R: 5′-TCGGAGGCAGTTCAAAGAG-3′ | ||||
| rs1065755 | Exon 6 | +8767 G>A | F: 5′-GCCTAAGCCTTCCCCAGGAG-3′ | HPYCH4IV |
| R: 5′-TGTAGAAGTAGTGCTCTGCCACACG-3′ | ||||
| rs1149045 | Intron 2 | −190 T>C | F: 5′-GCTACCCCTGGATTCTCCTC-3′ | HPYCH4IV |
| R: 5′-TAACCTGGCTCCCAAGTCTG-3′ | ||||
| rs1149046 | 5′-flanking | −546 C>T | F: 5′-GCTAAGAAGTGTTATTGCCAGGA-3′ | ApeKI |
| R: 5′-GGCCTGATGCAGAGTTGAAT-3′ | ||||
| rs3828051 | 5′-flanking | −944 T>C | F: 5′-CAGCAGTCGCTTTTTGTCTGTA-3′ | BccI |
| R: 5′-GTGGGAATGGTCGTTCTTGA-3′ | ||||
| rs1149048 | 5′-flanking | −1878 G>A | F: 5′-TGGAGGTGAACGAGGAGAAC-3′ | MspI |
| R: 5′-GAGCGGAGAAGTGACACAGA-3′ | ||||
| rs12404046 | 5′-flanking | −3023 A>T | F: 5′-AATCAAAGAGAGCTGGGCTTG-3′ | TfiI |
| R: 5′-GCATTCACTGCATTTCTTCCT-3′ |
The A of the translation initiation codon is denoted as +1.
Statistical analyses
Hardy–Weinberg equilibrium (HWE) was tested by a goodness-of-fit χ2 test with one degree of freedom. Chi-square tests were used to evaluate case–control difference for allele and genotype frequencies. The odds ratios (ORs) and their 95% confidence interval (CI) ranges were calculated. The best fitting model was selected by software PLINK34 (http://pngu.mgh.harvard.edu/pu rcell/plink/). Pair-wise linkage disequilibrium (LD) measures were estimated from the genotype data using the expectation–maximization (EM) algorithm implemented in HaploView version 4.0.35 One-way analysis of variance was used in the comparison of mean maximum Cobb angles with different genotypes in case-only analyses. We also performed permutation test (calculated by PLINK and HaploView) based on 10 000 permutations for multiple test adjustment. Significance was considered at P-value<0.05.
Results
Linkage disequilibrium structure and tagging SNP selection
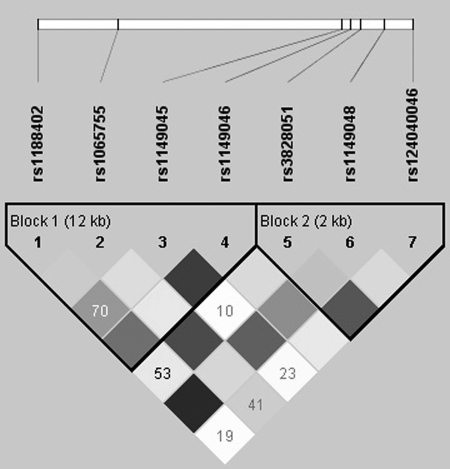
Using Hapmap Phase II data (release no. 21a/phase II, Jan07, population: CHB), we identified 17 common SNPs (MAF>0.10) in 17.89 kb region containing the MATN1 gene (including 5 kb upstream and 2 kb downstream of the gene). By LD analysis, two blocks were revealed by HaploView (Figure 1). In each block, four common (>2%) haplotypes were observed and accounted for at least 96% of all chromosomes in CHB population. Seven tSNPs (Figure 2) that predicted these common haplotypes were selected, four (SNP1: rs1188402, SNP2: rs1065755, SNP3: rs1149045 and SNP4: rs1149046) in block 1 and three (SNP5: rs3828051, SNP6: rs1149048 and SNP7: rs124040046) in block 2. The minimum R2 in each block was ≥0.80.
Figure 2.
Linkage disequilibrium (LD) structure of seven tag SNPs in 197 cases and 172 controls. The value within each diamond represents the pair-wise correlation between tag SNPs (measured as R2).
Case–control association studies
The genotype frequencies of the seven tSNPs are shown in Table 2. No significant deviation of genotype frequencies from the HWE was noted in either the control or the AIS group.
Table 2. Results of association analysis for seven tag SNPs in 197 cases and 172 controls.
| Genotypea | P-value (permutation P-valueb) | ||||
|---|---|---|---|---|---|
| SNPs | Case (n=197) | Control (n=172) | Allele | Genotyped | OR (95% CI)c |
| Block 1 | |||||
| rs1188402 | 56/91/49 | 41/86/45 | 0.38 (0.86) | 0.29 (0.78) | 1.15 (0.85–1.54) |
| rs1065755 | 141/47/9 | 128/40/4 | 0.31 (0.79) | 0.30 (0.79) | 0.81 (0.54–1.22) |
| rs1149045 | 60/92/45 | 58/89/25 | 0.13 (0.46) | 0.049 (0.18) | 0.80 (0.59–1.08) |
| rs1149046 | 85/83/28 | 75/84/13 | 0.33 (0.80) | 0.039 (0.14) | 0.86 (0.63–1.16) |
| Block 2 | |||||
| rs3828051 | 108/70/19 | 109/56/7 | 0.03 (0.15) | 0.03 (0.16) | 0.68 (0.49–0.96) |
| rs1149048 | 83/91/23 | 53/84/35 | 0.006 (0.034) | 0.006 (0.034e) | 1.52 (1.13–2.04) |
| rs12404046 | 130/58/9 | 131/37/4 | 0.03 (0.15) | 0.031 (0.15) | 0.64 (0.43–0.96) |
Significant results (permutation P-value <0.05) are shown in bold type.
The three values in the ‘genotype' column indicate the numbers of homozygotes with respect to the major allele, heterozygotes and homozygotes with respect to the minor allele, respectively.
Correction for multiple testing was performed using permutation test based on 10 000 permutations.
Calculated for the alleles.
The best-fitting model was selected by software PLINK (Cochran–Armitage trend test, genotypic test, dominant gene action test, recessive gene action test).
Permutation P-value was calculated by Cochran–Armitage trend test.
For the initial screening, we genotyped seven tSNPs in 197 cases and 172 controls. We examined association for allelic and genotypic differences between cases and controls. As showed in Table 2, three tSNPs in block 2 were found with significant associations (P<0.05), with the strongest association at rs1149048 (P=0.006). But after adjusting for multiple testing, only rs1149048 remained significant (permutation P-value=0.034, OR=1.52 within 95% CI=1.13–2.04).
Next, we validated the associated tSNPs (rs12404046, rs1149048 and rs3828051) in an additional sample of 222 cases and 288 controls. The pooled results of allele and genotype analysis are summarized in Table 3. We confirmed the significant association in rs1149048. In addition, another 290 controls were genotyped for rs1149048, and the results are also shown in Table 3. For SNP rs1149048, G allele had a higher risk for AIS than those carrying the A allele (P=0.0007, permutation P-value=0.0008, OR=1.35 within 95% CI=1.14–1.61). When comparing genotype of rs1149048, we found that GG conferred higher risk for AIS relative to AA and AG (P=0.005, OR=1.67 within 95% CI=1.18–2.38 for AA; P=0.001, OR=1.59 within 95% CI=1.22–2.08 for AG). Overall, individuals with genotype GG had a higher risk for AIS compared with AA+AG (P=0.0001, permutation P-value=0.0006, OR=1.61 within 95% CI=1.25–2.08, recessive model).
Table 3. Pooled results of analysis for three SNPs within block 2.
| Genotypea | P-value (permutation P-valueb) | ||||
|---|---|---|---|---|---|
| SNPs | Case (n=419) | Control (n=460) | Allele | Genotyped | OR (95% CI)c |
| rs3828051 | 228/158/32 | 278/159/23 | 0.040 (0.128) | 0.042 (0.12) | 0.79 (0.64–0.99) |
| rs1149048e | 174/180/65 | 141/233/86 | 0.0027 (0.014) | 0.0008 (0.0034f) | 1.34 (1.11–1.62) |
| rs12404046 | 279/119/20 | 334/113/13 | 0.028 (0.095) | 0.027 (0.10) | 0.76 (0.59–0.97) |
Significant results (permutation P-value<0.05) are shown in bold type.
The three values in the ‘genotype' column indicate the numbers of homozygotes with respect to the major allele, heterozygotes and homozygotes with respect to the minor allele, respectively.
Correction for multiple testing was performed using permutation test based on 10 000 permutations.
Calculated for the alleles.
The best-fitting model was selected by software PLINK (Cochran–Armitage trend test, genotypic test, dominant gene action test, recessive gene action test).
Control sample size was increased to 750 to confirm the significant association. The genotype frequencies were (N=750) 231, 376 and 143 for AA, AG and GG, respectively. Both genotypic and allelic associations were significant (P=0.0006, 0.0008 by permutation).
Permutation P-value was calculated by recessive gene action test.
To perform haplotype-based association analyses, we re-examined LD structures within all genotype data for a single-SNP association analysis. The extent and patterns of LD essentially were identical to those obtained from Hapmap (Figure 2). There were two blocks each with four common haplotypes, which covered more than 96% of all observed haplotypes. When the estimated haplotype frequencies were compared between cases and controls, the AAA haplotype was more prevalent in the control group, whereas the GGT haplotype was more common among patients (Table 4).
Table 4. Associations between rs3828051, rs1149048 and rs12404046 haplotypes and disease risk in total 419 cases and 460 controls.
| Haplotypesa | Cases (n=419) | Controls (n=460) | P-valueb | OR (95% CI)c |
|---|---|---|---|---|
| AAA | 310.0 (0.369) | 405.0 (0.440) | 0.0027 | 1.34 (1.11–1.62) |
| AGA | 306.7 (0.365) | 310.0 (0.337) | 0.217 | 1.13 (0.93–1.38) |
| GGT | 160.7 (0.191) | 139.0 (0.151) | 0.0249 | 0.75 (0.58–0.96) |
| GGA | 62.6 (0.075) | 66.0 (0.072) | 0.8192 | 1.05 (0.73–1.50) |
The position of rs1149048 is underlined in the haplotypes.
Haplotypes observed with <0.02 in both controls and cases have been dropped.
P-values were calculated using χ2 test.
Odds ratios (ORs); 95% confidence intervals (CIs) are shown in parentheses.
We examined whether confounding effects, such as age and sex, affected the association results and found no relationship between the genotype and these factors (Table 5). The association was strongly significant in the female group but negative in the male group, which may be due to the small sample size for male cases and controls (Table 6).
Table 5. Correlation between age and genotype at rs1149048 in MATN1.
| Mean±SD age (in years) for genotype | ||||
|---|---|---|---|---|
| Population | AA | AG | GG | P-valuea |
| Case | 15.15±2.58 | 15.26±2.90 | 15.77±3.17 | 0.296 |
| Control | 16.07±0.91 | 16.21±1.10 | 16.085±0.93 | 0.256 |
P-value was calculated using the Kruskal–Wallis test.
Table 6. Genotype at rs1149048 in MATN1, stratified by sex.
| Male | Female | |||
|---|---|---|---|---|
| Measure | Case | Control | Case | Control |
| No. of subjects | ||||
| ALL | 57 | 47 | 362 | 413 |
| GG | 27 | 20 | 147 | 121 |
| AG | 21 | 18 | 159 | 215 |
| AA | 9 | 9 | 56 | 77 |
| Allele frequency (%) | 0.34 | 0.38 | 0.37 | 0.44 |
| P-valuea | 0.60 | 0.0044 | ||
P-value for allelic difference between the patients and the control groups for each sex determined by the χ2 test.
Case-only association studies
A subgroup of AIS patients (N=267) who has not been braced and received continued observation or surgery was included in a case-only study to determine the effect of variations on disease severity. We found that only rs1149048 polymorphism was associated with curve severity. The mean maximal Cobb angle of patients with the high-risk genotype GG (37.91±17.08°) was higher than that of those with genotype AA (33.88±14.68°) and AG (32.25±12.42°), which were statistically significant differences (P=0.002 by ANOVA test). Overall, individuals with genotype GG tended to develop a larger Cobb angle than those with genotype AA+AG (P=0.002, recessive model) (Table 7).
Table 7. Association between curve severity and genotype of rs1149048 was examined among the cases to determine if the SNP also correlated with disease severity.
| Genotype (number of cases) | Maximum Cobb angle (°) mean±SD | P-value |
|---|---|---|
| AA (38) | 32.11±11.67 | 0.034a |
| AG (115) | 32.25±12.42 | 0.003b |
| GG (114) | 37.91±17.08 | Reference |
The difference among AA, AG and GG (P=0.002) compared using one-way ANOVA test.
The difference among AA, AG and GG (P=0.03) compared using Kruskal–Wallis test.
The difference between AA and GG compared using LSD test.
The difference between AG and GG compared using LSD test.
Discussion
In this study, using candidate gene association study, we identified that rs1149048 polymorphism in MATN1 promoter region was associated with both susceptibility and disease progression in AIS. The results showed that MATN1 was both a disease predisposition and progression gene. We confirmed the earlier finding of association between AIS and MATN1 using a small sample of 81 Caucasian trios by an STRP marker.15 It appears that the effect may be present across ethnic groups. Also, it indicated that MATN1 was an important gene that involved in the complex genetic etiopathogenesis of AIS. Furthermore, this study provided evidence to support that rs1149048 or other SNPs in LD with this SNP in the promoter of MATN1 may account for disease predisposition.
The candidate gene association study has been proved an efficient tool to study complex diseases. Linkage and association approaches are commonly used to search for disease predisposition genes. Although valuable for detecting genetic loci in single-gene disorders, linkage analysis of common diseases is less powerful than association study in the analysis of complex trait. On the other hand, an association study always tends to produce spurious results due to population stratification, effect heterogeneity and type I errors. Therefore, replications are usually necessary for the confirmation of an association finding. Unfortunately, it is reported that only 16–30%29 of the putative association findings could be successfully replicated. Benjamin et al29 suggested that it was difficult to replicate association findings at the SNP or haplotype level and proposed a gene-based approach in which all common variations within a candidate gene are considered jointly. By using tSNP approach, it is possible to study all but rare genetic variants or haplotypes by a small set of relatively common SNPs. Empirical data have demonstrated that tSNPs selected from the HapMap population samples can effectively capture common variation and provide good power to detect an association under a disease model of modest risk.13 On the basis of the latest Hapmap database (release no. 21a/phase II, Jan 07), our comprehensive association analysis consisted of seven informative tSNPs to provide a good coverage of variants in MATN1. Therefore, this approach is more powerful and sensitive than the earlier study using one STRP marker.15
The earlier study15 suggested that idiopathic scoliosis is associated with MATN1 in a TDT study of 81 trios and detected an association with one allele of microsatellite in 3′-UTR of the gene. Here we further explore this gene for association with AIS by using a set of tSNPs and a larger sample size. In our gene-based replication study, we used the stepwise method to explore the associated tag SNPs. In the first initial screening stage, we found three SNPs with significant associations in block 2. It was then followed by the second validation stage in another sample that confirmed the results of the initial stage. The G allele of rs1149048 predisposed to AIS. However, this study confirmed a different marker other than the marker found by Montanaro et al,15 which was only a microsatellite marker. Also, the STRP reported earlier is most likely representing or in LD with another causative polymorphism. Therefore, our study supplemented in detail on top of the paper by Montrnaro15 et al.
In addition, the effect of gene polymorphisms on curve severity of AIS was also analyzed in a subgroup of AIS patients who received continued observation or surgery. The possible role of MATN1 polymorphisms in disease progression was assessed quantitatively with the maximum Cobb angle. We found that rs1149048 polymorphism was associated with curve severity. Individuals with GG genotype had a larger mean maximal Cobb angle than those with genotype AA+AG, indicating that the G allele of rs1149048 had risk effect on risk of AIS under a recessive model that was consistent with case–control association study. The results showed that MATN1 gene is also a disease modifier gene of AIS. In the literature, there are only two genes to be considered as disease modifier genes of AIS, including ESR118 and IGF1.36 Genetic analysis of disease modifier gene might be applicable to the prediction of progression of scoliosis and could be helpful in early clinical investigation and treatment.
A recent analysis37, 38 of the major regulatory regions of chicken matrilin-1 gene in transgenic mice revealed a distal promoter region between −2011 and −338 that includes cartilage-specific control element. By sequence alignment among various vertebrate species, Rentsendorj38 identified four conserved sequence blocks (distal promoter element (Dpe)1, Dpe2, Pe1 and Ine) in the promoters of amniote matrillin-1 gene. Interestingly, SNP rs1149048 in our study is located in the distal promoter region of MATN1, 126 bp downstream of the Dpe2. It is plausible that the function of rs1149048 could be mediated through an interaction with one of these conserved regulatory blocks like Dpe2 or other SNPs in LD with it.
Several limitations of this study should be considered. First, we are not certain if rs1149048 is a specific causal SNP or just a marker in LD with another polymorphism that plays a causative role. Therefore, further fine mapping of the haploblocks encompassing the region close to rs1149048 polymorphism will be necessary to clarify the susceptible allele of AIS. Furthermore, the functional analysis of these polymorphisms may add further information on genetic association studies and elucidate the role of different genotypes in the pathogenesis of AIS. Second, ethnic differences may account for differences in disease susceptibility. Our sample is entirely from Chinese population. The finding may not be generalized to other ethnic groups. Further evaluation from other ethnic populations might help clarify the real genetic effect.
In conclusion, this study provides evidence that the tSNP rs1149048 in the MATN1 promoter region was associated with both susceptibility and curve severity to AIS. The results suggested that MATN1 was both an AIS predisposition and progression gene.
Acknowledgments
We thank Dr Bing Wang, Dr Yang Yu, Dr Zezhang Zhu, Dr Feng Zhu, Dr Bangping Qian and Dr Weiwei Ma for their assistance. This study was supported by grants from the National Natural Science Foundation of China (30672131).
References
- Kane WJ. Scoliosis prevalence: a call for a statement of terms. Clin Orthop Relat Res. 1977;126:43–46. [PubMed] [Google Scholar]
- Lowe TG, Edgar M, Margulies JY, et al. Etiology of idiopathic scoliosis: current trends in research. J Bone Joint Surg Am. 2000;82:1157–1168. doi: 10.2106/00004623-200008000-00014. [DOI] [PubMed] [Google Scholar]
- Ogilvie JW, Braun J, Argyle V, Nelson L, Meade M, Ward K. The search for idiopathic scoliosis genes. Spine. 2006;31:679–681. doi: 10.1097/01.brs.0000202527.25356.90. [DOI] [PubMed] [Google Scholar]
- Miller NH. Genetics of familial idiopathic scoliosis. Clin Orthop Relat Res. 2002;401:60–64. doi: 10.1097/00003086-200208000-00009. [DOI] [PubMed] [Google Scholar]
- Justice CM, Miller NH, Marosy B, Zhang J, Wilson AF. Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine. 2003;28:589–594. doi: 10.1097/01.BRS.0000049940.39801.E6. [DOI] [PubMed] [Google Scholar]
- Wise CA, Barnes R, Gillum J, Herring JA, Bowcock AM, Lovett M. Localization of susceptibility to familial idiopathic scoliosis. Spine. 2000;25:2372–2380. doi: 10.1097/00007632-200009150-00017. [DOI] [PubMed] [Google Scholar]
- Salehi LB, Mangino M, De Serio S, et al. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum Genet. 2002;111:401–404. doi: 10.1007/s00439-002-0785-4. [DOI] [PubMed] [Google Scholar]
- Alden KJ, Marosy B, Nzegwu N, Justice CM, Wilson AF, Miller NH. Idiopathic scoliosis: identification of candidate regions on chromosome 19p13. Spine. 2006;31:1815–1819. doi: 10.1097/01.brs.0000227264.23603.dc. [DOI] [PubMed] [Google Scholar]
- Chan V, Fong GC, Luk KD, et al. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. 2002;71:401–406. doi: 10.1086/341607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiardes S, Veile R, Allen M, et al. SNTG1, the gene encoding gamma1-syntrophin: a candidate gene for idiopathic scoliosis. Hum Genet. 2004;115:81–89. doi: 10.1007/s00439-004-1121-y. [DOI] [PubMed] [Google Scholar]
- Ocaka L, Zhao C, Reed JA, et al. Assignment of two loci for autosomal dominant Adolescent Idiopathic Scoliosis (AIS) to chromosomes 9q31.2–q34.2 and 17q25.3–qtel. J Med Genet. 2008;45:87–92. doi: 10.1136/jmg.2007.051896. [DOI] [PubMed] [Google Scholar]
- Miller NH, Justice CM, Marosy B, et al. Identification of candidate regions for familial idiopathic scoliosis. Spine. 2005;30:1181–1187. doi: 10.1097/01.brs.0000162282.46160.0a. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Tang NL, Yeung HY, Miller N. Genetic association of complex traits: using idiopathic scoliosis as an example. Clin Orthop Relat Res. 2007;462:38–44. doi: 10.1097/BLO.0b013e3180d09dcc. [DOI] [PubMed] [Google Scholar]
- Wu J, Qiu Y, Zhang L, Sun Q, Qiu X, He Y. Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine. 2006;31:1131–1136. doi: 10.1097/01.brs.0000216603.91330.6f. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Parisini P, Greggi T, et al. Evidence of a linkage between matrilin-1 gene (MATN1) and idiopathic scoliosis. Scoliosis. 2006;1:21. doi: 10.1186/1748-7161-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Gordon D, Zhang D, et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007;80:957–965. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XS, Tang NL, Yeung HY, et al. Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. Spine. 2007;32:1748–1753. doi: 10.1097/BRS.0b013e3180b9f0ff. [DOI] [PubMed] [Google Scholar]
- Inoue M, Minami S, Nakata Y, et al. Association between estrogen receptor gene polymorphisms and curve severity of idiopathic scoliosis. Spine. 2002;27:2357–2362. doi: 10.1097/00007632-200211010-00009. [DOI] [PubMed] [Google Scholar]
- Carr AJ, Ogilvie DJ, Wordsworth BP, Priestly LM, Smith R, Sykes B. Segregation of structural collagen genes in adolescent idiopathic scoliosis. Clin Orthop Relat Res. 1992;274:305–310. [PubMed] [Google Scholar]
- Miller NH, Mims B, Child A, Milewicz DM, Sponseller P, Blanton SH. Genetic analysis of structural elastic fiber and collagen genes in familial adolescent idiopathic scoliosis. J Orthop Res. 1996;14:994–999. doi: 10.1002/jor.1100140621. [DOI] [PubMed] [Google Scholar]
- Marosy B, Justice CM, Nzegwu N, Kumar G, Wilson AF, Miller NH. Lack of association between the aggrecan gene and familial idiopathic scoliosis. Spine. 2006;31:1420–1425. doi: 10.1097/01.brs.0000219944.18223.52. [DOI] [PubMed] [Google Scholar]
- Zorkol'tseva IV, Liubinskii OA, Sharipov RN, Zaidman AM, Aksenovich TI, Dymshits GM. Analysis of polymorphism of the number of tandem repeats in the aggrecan gene exon G3 in the families with idiopathic scoliosis. Genetika. 2002;38:259–263. [PubMed] [Google Scholar]
- Chen Q, Johnson D, Goetinck PF. Assembly of cartilage matrix protein filamentous networks. Mol Biol Cell. 1995;6:2221. doi: 10.1091/mbc.10.7.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Johnson DM, Haudenschild DR, Goetinck PF. Progression and recapitulation of the chondrocyte differentiation program: cartilage matrix protein is a marker for cartilage maturation. Dev Biol. 1995;172:293–306. doi: 10.1006/dbio.1995.0024. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- Chapman KL, Mortier GR, Chapman K, Loughlin J, Grant ME, Briggs MD. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat Genet. 2001;28:393–396. doi: 10.1038/ng573. [DOI] [PubMed] [Google Scholar]
- Blank RD, Raggio CL, Giampietro PF, Camacho NP. A genomic approach to scoliosis pathogenesis. Lupus. 1999;8:356–360. doi: 10.1177/096120339900800505. [DOI] [PubMed] [Google Scholar]
- Giampietro PF, Raggio CL, Blank RD. Synteny-defined candidate genes for congenital and idiopathic scoliosis. Am J Med Genet. 1999;83:164–177. [PubMed] [Google Scholar]
- Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Stram DO. Tag SNP selection for association studies. Genet Epidemiol. 2004;27:365–374. doi: 10.1002/gepi.20028. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Yeung HY, Tang NL, Lee KM, et al. Genetic association study of insulin-like growth factor-I (IGF-I) gene with curve severity and osteopenia in adolescent idiopathic scoliosis. Stud Health Technol Inform. 2006;123:18–24. [PubMed] [Google Scholar]
- Karcagi I, Rauch T, Hiripi L, et al. Functional analysis of the regulatory regions of the matrilin-1 gene in transgenic mice reveals modular arrangement of tissue-specific control elements. Matrix Biol. 2004;22:605–618. doi: 10.1016/j.matbio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Rentsendorj O, Nagy A, Sinkó I, Daraba A, Barta E, Kiss I. Highly conserved proximal promoter element harbouring paired Sox9-binding sites contributes to the tissue- and developmental stage-specific activity of the matrilin-1 gene. Biochem J. 2005;389:705–716. doi: 10.1042/BJ20050214. [DOI] [PMC free article] [PubMed] [Google Scholar]