Abstract
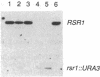
Genes CDC24, CDC42, and CDC43 are required for the establishment of cell polarity and the localization of secretion in Saccharomyces cerevisiae; mutants defective in these genes fail to form buds and display isotropic expansion of the cell surface. To identify other genes that may be involved in these processes, we screened yeast genomic DNA libraries for heterologous genes that, when overexpressed from a plasmid, can suppress a temperature-sensitive cdc24 mutation. We identified four such genes. One of these proved to be CDC42, which has previously been shown to be a member of the rho (ras-homologous) family of genes, and a second is a newly identified ras-related gene that we named RSR1. RSR1 maps between CDC62 and ADE3 on the right arm of chromosome VII; its predicted product is approximately 50% identical to other proteins in the ras family. Deletion of RSR1 is nonlethal but disrupts the normal pattern of bud site selection. Although both CDC42 and RSR1 can suppress cdc24 and both appear to encode GTP-binding proteins, these genes do not themselves appear to be functionally interchangeable. However, one of the other genes that was isolated by virtue of its ability to suppress cdc24 can also suppress cdc42. This gene, named MSB1, maps between ADE9 and HIS3 on the right arm of chromosome XV.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989 Mar;121(3):463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. M., Wang M. D., Vartak N. B., Liu L. Acquisition of new metabolic capabilities: multicopy suppression by cloned transaminase genes in Escherichia coli K-12. Gene. 1988 May 30;65(2):195–202. doi: 10.1016/0378-1119(88)90456-8. [DOI] [PubMed] [Google Scholar]
- Booher R., Beach D. Interaction between cdc13+ and cdc2+ in the control of mitosis in fission yeast; dissociation of the G1 and G2 roles of the cdc2+ protein kinase. EMBO J. 1987 Nov;6(11):3441–3447. doi: 10.1002/j.1460-2075.1987.tb02667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Capon D. J., Chen E. Y., Levinson A. D., Seeburg P. H., Goeddel D. V. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983 Mar 3;302(5903):33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Clarke S., Vogel J. P., Deschenes R. J., Stock J. Posttranslational modification of the Ha-ras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K. G., Steensma H. Y., Kaback D. B., Pringle J. R. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and characterization of the CDC24 gene and adjacent regions of the chromosome. Mol Cell Biol. 1986 Dec;6(12):4516–4525. doi: 10.1128/mcb.6.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der C. J., Finkel T., Cooper G. M. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986 Jan 17;44(1):167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell. 1987 Sep 25;50(7):1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Drabkin H. J., RajBhandary U. L. Attempted expression of a human initiator tRNA gene in Saccharomyces cerevisiae. J Biol Chem. 1985 May 10;260(9):5596–5602. [PubMed] [Google Scholar]
- Field C., Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980 Jul;86(1):123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAWTHORNE D. C., FRIIS J. OSMOTIC-REMEDIAL MUTANTS. A NEW CLASSIFICATION FOR NUTRITIONAL MUTANTS IN YEAST. Genetics. 1964 Nov;50:829–839. doi: 10.1093/genetics/50.5.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Pringle J. R., Reid B. J. Genetic control of the cell division cycle in yeast. Science. 1974 Jan 11;183(4120):46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hayles J., Aves S., Nurse P. suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J. 1986 Dec 1;5(12):3373–3379. doi: 10.1002/j.1460-2075.1986.tb04653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Barker D. G., Nurse P. Cloning and characterization of the Schizosaccharomyces pombe DNA ligase gene CDC17. Gene. 1986;41(2-3):321–325. doi: 10.1016/0378-1119(86)90114-9. [DOI] [PubMed] [Google Scholar]
- Lawrence C. B., Goldman D. A., Hood R. T. Optimized homology searches of the gene and protein sequence data banks. Bull Math Biol. 1986;48(5-6):569–583. doi: 10.1007/BF02462324. [DOI] [PubMed] [Google Scholar]
- MacKay V. L. Cloning of yeast STE genes in 2 microns vectors. Methods Enzymol. 1983;101:325–343. doi: 10.1016/0076-6879(83)01025-3. [DOI] [PubMed] [Google Scholar]
- Madaule P., Axel R., Myers A. M. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Feb;84(3):779–783. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Ohya Y., Ohsumi Y., Anraku Y. Nucleotide sequence of the CLS4 (CDC24) gene of Saccharomyces cerevisiae. Gene. 1987;54(1):125–132. doi: 10.1016/0378-1119(87)90354-4. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic Mapping in Saccharomyces IV. Mapping of Temperature-Sensitive Genes and Use of Disomic Strains in Localizing Genes. Genetics. 1973 May;74(1):33–54. doi: 10.1093/genetics/74.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., Contopoulou C. R., Kans J. A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast. 1989 Sep-Oct;5(5):321–403. doi: 10.1002/yea.320050503. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Miyamoto S., Ohsumi Y., Anraku Y. Calcium-sensitive cls4 mutant of Saccharomyces cerevisiae with a defect in bud formation. J Bacteriol. 1986 Jan;165(1):28–33. doi: 10.1128/jb.165.1.28-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V., Chardin P., Lerosey I., Olofsson B., Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the 'effector' region. Oncogene. 1988 Aug;3(2):201–204. [PubMed] [Google Scholar]
- Powers S., Kataoka T., Fasano O., Goldfarb M., Strathern J., Broach J., Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell. 1984 Mar;36(3):607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987 May 22;49(4):527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Harold F. M. Dual roles for calcium ions in apical growth of Neurospora crassa. J Gen Microbiol. 1988 Sep;134(9):2623–2631. doi: 10.1099/00221287-134-9-2623. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat B. F., Adams A., Pringle J. R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981 Jun;89(3):395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat B. F., Pringle J. R. A mutant of yeast defective in cellular morphogenesis. Science. 1978 Jun 9;200(4346):1171–1173. doi: 10.1126/science.349694. [DOI] [PubMed] [Google Scholar]
- Wilkinson L. E., Pringle J. R. Transient G1 arrest of S. cerevisiae cells of mating type alpha by a factor produced by cells of mating type a. Exp Cell Res. 1974 Nov;89(1):175–187. doi: 10.1016/0014-4827(74)90200-6. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Papageorge A. G., Kung H. F., Bekesi E., Robins T., Johnsen M., Vass W. C., Lowy D. R. Mutational analysis of a ras catalytic domain. Mol Cell Biol. 1986 Jul;6(7):2646–2654. doi: 10.1128/mcb.6.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]