Abstract
Type III hyperlipoproteinemia (HLP) is mainly found in homozygous apolipoprotein (APO) E2 (R158C) carriers. Genetic factors contributing to the expression of type III HLP were investigated in 113 hyper- and 52 normolipidemic E2/2 subjects, by testing for polymorphisms in APOC3, APOA5, HL (hepatic lipase) and LPL (lipoprotein lipase) genes. In addition, 188 normolipidemic Dutch control panels (NDCP) and 141 hypertriglyceridemic (HTG) patients were genotyped as well. No associations were found for four HL gene polymorphisms and two LPL gene polymorphisms and type III HLP. The frequency of the rare allele of APOC3 3238 G>C and APOA5 −1131 T>C (in linkage disequilibrium) was significantly higher in type III HLP patients when compared with normolipidemic E2/2 subjects, 15.6 vs 6.9% and 15.1 vs 5.8%, respectively, (P<0.05). Furthermore, the frequencies of the APOA5 c.56 G>C polymorphism and LPL c.27 G>A mutation were higher in type III HLP patients, though not significant. Some 58% of the type III HLP patients carried either the APOA5 −1131 T>C, c.56 G>C and/or LPL c.27 G>A mutation as compared to 27% of the normolipidemic APOE2/2 subjects (odds ratio 3.7, 95% confidence interval=1.8–7.5, P<0.0001). The HTG patients showed similar allele frequencies of the APOA5, APOC3 and LPL polymorphisms, whereas the NDCP showed similar allele frequencies as the normolipidemic APOE2/2. Patients with the APOC3 3238 G>C/APOA5 −1131 T>C polymorphism showed a more severe hyperlipidemia than patients without this polymorphism. Polymorphisms in lipolysis genes associate with the expression and severity of type III HLP in APOE2/2.
Keywords: type III hyperlipoproteinemia, apoAV, apoCIII, lipoprotein lipase, SNP analysis
Introduction
Patients with type III hyperlipoproteinemia (HLP) are characterized by elevated levels of total cholesterol and triglycerides (TGs) due to high plasma levels of chylomicron and very low-density lipoprotein (VLDL) remnants enriched in cholesterol esters and apolipoprotein E (APOE).1, 2
APOE, a major constituent of chylomicron and VLDL remnants, serves as a ligand for the receptor-mediated uptake of these particles by the liver.3 In type III HLP, APOE mutations lead to an impaired clearance of remnant lipoproteins by hepatic lipoprotein receptors. There are three common genetic variants of APOE: APOE2 (Arg158 → Cys), APOE3 (Cys112; Arg158) and APOE4 (Cys112 → Arg). These isoforms are encoded by three co-dominant alleles that are located at one single gene locus on chromosome 19. In comparison to the other two isoforms, APOE2 has less than 1% binding capacity for the hepatic LDL receptor.4 Most type III HLP patients (>90%) are homozygous carriers of APOE2 (Arg158 → Cys).4 In Caucasian populations, APOE2 homozygosity occurs with a frequency of about 1%, whereas the frequency of type III HLP is about 1–7 per 5000.5, 6 A minority of the APOE2 homozygous subjects will develop type III HLP, indicating that type III HLP is a multifactorial disorder requiring additional genetic and environmental factors for its clinical manifestation.4, 7, 8 It has been suggested that contributors to the expression of type III HLP include factors causing (1) an overproduction of lipoproteins, (2) an impaired lipolysis of lipoproteins or (3) an impaired hepatic uptake of remnants.4, 9 Insulin resistance is associated with high TG levels caused by an increased VLDL production.10, 11 Earlier we found an association of high insulin levels with the expression of type III HLP.6, 12
Several groups studied genetic factors that may contribute to the expression of type III HLP.13, 14, 15, 16 However, in the present study, we have studied a larger type III HLP cohort. In addition, we used a normolipidemic APOE2 homozygote cohort as control group. Mutations in genes involved in lipolytic conversion, such as LPL (lipoprotein lipase), HL (hepatic lipase) and APOC3 have been associated with hyperlipidemia (for reviews, see references17, 18, 19, 20). In addition, Zhang et al13 observed an increased allele frequency for the LPL N291S mutation in type III HLP patients when compared with the general population. Single nucleotide polymorphisms (SNPs) in the APOA5 gene (11q23) were found to be strongly associated with plasma TG levels.21, 22
Data from family studies on type III HLP indicate that one or more genes are possible additional genetic factors predisposing to type III HLP.4 However, from these studies it was not evident which additional genes were involved or whether the study population was very small. In the present study, a significant population of both normolipidemic and hyperlipidemic E2/2 subjects was collected to determine additional genetic risk factors contributing to the expression of type III HLP in APOE2 homozygotes. For comparison, these genetic risk factors were also typed in hypertriglyceridemic (HTG) patients23, 24 – they partly match their elevated VLDL/TG phenotype with type III HLP patients – and a normolipidemic Dutch control panel (NDCP).
Methods
Subjects
The study population consisted of 167 unrelated homozygous carriers of APOE2 (Arg158 → Cys). Type III HLP patients were defined as having total cholesterol and TG levels ≥90th percentile, VLDL cholesterol/TG ratio >0.3 (mg/100 ml/mg/100 ml) and/or VLDL cholesterol/VLDL TG ratio of >0.8 (mmol/l/mmol/l), whereas normolipidemic E2/2 subjects had total cholesterol and TG levels <90th percentile according to the age- and sex-related percentile levels of the Prospective Cardiovascular Münster Study (PROCAM).25 Ultracentrifuge data were not available from 24 HLP type III patients. Two HLP APOE2 homozygotes were excluded from the analysis because their plasma TG levels were normal.
In total, 102 type III HLP patients and 9 normolipidemic E2/2 subjects were recruited from the outpatient Lipid Clinics of the University Medical Centers of Leiden, Nijmegen, Amsterdam and Rotterdam. Ten unrelated carriers (two type III HLP patients and eight normolipidemic E2/2 subjects) were detected during a population-based study among 2018 randomly selected 35-year-old men.26 In total, 44 E2/2 subjects (9 type III HLP patients and 35 normolipidemic E2/2 subjects) were collected from the Rotterdam Study, a prospective cohort study of 6870 healthy persons aged 55 years and older, investigating determinants of chronic diseases.27
In this study, we included 113 type III HLP patients and 52 normolipidemic E2/2 subjects. Clinical data and blood samples from type III HLP patients were collected before lipid-lowering medication. One normolipidemic E2/2 subject and one type III HLP patient with a history of pancreatitis were not included in the study.
The selection of the 141 HTG patients was described earlier.23 The 188 NDCPs were selected on APOE genotype, and total plasma cholesterol, TGs and high-density lipoprotein (HDL) levels, between the 25th and 75th percentile. Informed consent was given by each participant and the study was approved by the Ethics Committee of our hospital.
Vascular disease was defined as the presence of coronary artery disease (angina pectoris, 70% stenosis on coronary arteriography, myocardial infarction, coronary by-pass or percutaneous transluminal coronary angioplasty) and/or cerebrovascular disease (stroke or transient ischemic attack) and/or peripheral vascular disease. Hypertension was defined as systolic blood pressure of ≥160 mm Hg, or diastolic blood pressure of ≥90 mm Hg, or the use of medication for hypertension.28 Less than 20% of the type III HLP patients were treated with antihypertensive medication: 11% with a betablocker and/or diuretic and 8% with other antihypertensive drugs. About 6% of the normolipidemic subjects were treated with antihypertensive medication. Four patients received antidiabetic medication.
Hyperinsulinemia was defined as fasting insulin concentrations ≥100 pmol/l. The diagnostic criterion for diabetes mellitus was fasting blood glucose ≥7 mmol/l. Smoking was defined as the consumption of at least 10 cigarettes per day. The non-smokers also included ex-smokers, who stopped smoking for at least 1 year. Alcohol consumers were defined as subjects with alcohol consumption of 2 or more grams per day.
APOE phenotyping and genotyping
APOE phenotypes were determined by isoelectric focusing of delipidated serum samples and after cysteamine treatment followed by immunoblotting with a polyclonal anti-APOE antiserum as described.29 The results were confirmed by APOE genotyping.30
Lipid and lipoprotein analysis
With the exception of the sampling of 38 E2/2 subjects originating from the Rotterdam Study, all blood samples were collected after an overnight fast. Serum was obtained after centrifugation at 1500 g for 15 min. Total serum cholesterol and TG levels were measured with commercially available kits. Serum HDL cholesterol concentration was measured after precipitation of VLDL and LDL with phosphotungstic acid and MgCl2.31 Serum (3 ml) was ultracentrifuged for 15 h at 232 000 g at 15°C in a TL-100 tabletop ultracentrifuge. The ultracentrifugate was divided into a density (d) <1.006 and 1.006 <d<1.25 g/ml fractions, designated as the VLDL and IDL+LDL−HDL fractions, respectively.
Serum insulin and glucose measurements
Insulin and glucose concentrations were measured only in fasting blood samples. The insulin concentration was determined by a radioimmunoassay (Ins-Ria-100; MedGenix). The antibody of this assay cross-reacts with proinsulin (40%) but not with C-peptide (<0.001%).32 Serum glucose was determined by the automated hexokinase method of Hitachi 747, Boehringer Mannheim-Hitachi.
DNA analyses
Genomic DNA was isolated from leukocytes.33 The following mutations or polymorphisms were identified with PCR followed by restriction enzyme analysis as described earlier: the 3238 G>C (Sst-1; rs5128)34 polymorphism in 3′-untranslated region (UTR) of exon 4 of the APOC3 gene, LPL c.27 G>A (D9N; rs1801177), LPL c.1342 C>G (S447X; rs328),35 LPL c.874 A>G (N291S; rs268),13 HL c.219 G>A (V73M; rs6078), HL –480 C>T (rs8192701),36 HL c.1005 A>G (L334F; rs3829462)37 and HL c.609 C>G (T202T; rs6084)38 in the LPL and HL genes, respectively. Analysis of both the APOA5 polymorphisms −1131 T>C (SNP3; rs662799) in the APOA5 promoter region and the c.56 G>C (S19W, rs3135506) in exon 3 of the APOA5 gene was as described earlier.23
Statistical analyses
Differences between groups were tested with the χ2 test for dichotomous and categorical variables and the unpaired Student's t-test for continuous variables. As total cholesterol, total TG and plasma insulin levels showed non-Gaussian distributions, these parameters were logarithmically transformed before analysis. Untransformed levels are shown in the tables.
For each polymorphism or mutation, the Hardy–Weinberg equilibrium was calculated using the gene-counting method and differences were assessed by the χ2 test. The χ2 test or Fisher's exact test were applied to compare genotype and allele frequencies between type III HLP patients, normolipidemic E2/2 subjects, NDCP and HTG patients. A logistic regression model was used to examine the association between the presence of a mutation and the occurrence of type III HLP. The strength of the association was estimated as the odds ratio (OR) with 95% confidence intervals (CIs). P-values lower than 0.05 were considered significant. Selection of all SNPs in this study was based on literature data, describing association of these SNPs with lipid phenotype. Therefore adjustment for multiple testing was not applied (replication). Statistical analyses were performed with SPSS statistical software (version 14.01; SPSS, Chicago, IL, USA). Linkage disequilibrium (LD) estimations in the APOC3/A5 region were estimated using Haploview.3.33.39
Results
Baseline characteristics of the study populations
The mean age of type III HLP patients was lower and their BMI was higher compared with normolipidemic E2/2 subjects (Table 1). The prevalence of vascular disease was increased in type III HLP patients, whereas no differences in the occurrence of hypertension, diabetes mellitus and the number of smokers and alcohol consumers were found between the groups. Furthermore, type III HLP patients had significantly higher total TG and insulin levels as compared with normolipidemic E2/2 subjects, whereas HDL cholesterol was decreased. Characteristics of the normolipidemic panel (NDCP) and HTG patients are summarized in Table 1.
Table 1. Clinical and biochemical characteristics of type III HLP patients, normolipidemic E2/2 subjects, normal Dutch controls and hypertriglyceridemic patients.
| Type III HLP | Normo | NDCP | HTG | |||||
|---|---|---|---|---|---|---|---|---|
| N | N | N | N | |||||
| Men | 76 (67%) | 113 | 32 (61%) | 52 | 90 (48%) | 188 | 119 (84%) | 141 |
| Women | 37 (33%) | 113 | 20 (39%) | 52 | 97 (52%) | 188 | 22 (16%) | 141 |
| Age mean (years) | 50.4** | 113 | 64.0 | 52 | 41 | 187 | 53 | 141 |
| Age range (years) | 27–80 | 113 | 27–80 | 52 | 6–78 | 187 | 26–78 | 141 |
| Body mass index (kg/m2) | 26.9±3.8* | 113 | 25.3±4.0 | 49 | — | — | — | — |
| Vascular disease | 46 (41%)* | 113 | 7 (15%) | 39 | — | — | — | — |
| Hypertension | 31 (27%) | 113 | 14 (26%) | 42 | — | — | — | — |
| Diabetes mellitus | 11 (10%) | 113 | 5 (10%) | 40 | — | — | — | — |
| Smokers | 45 (40%) | 112 | 19 (39%) | 43 | — | — | — | — |
| Alcohol consumers | 78 (75%) | 105 | 29 (78%) | 30 | — | — | — | — |
| Total cholesterol (mmol/l) | 11.0±3.5a,** | 113 | 5.7±1.0 | 52 | 5.1±0.72a | 188 | 8.8±3.89 | 141 |
| Total triglycerides (mmol/l) | 6.7±4.7a,** | 113 | 2.0±0.7 | 47 | 1.1±0.35a | 188 | 14.3±13.80a | 137 |
| HDL-C (mmol/l) | 1.0±0.3** | 113 | 1.4±0.3 | 48 | 1.3±0.23a | 188 | 0.7±0.20 | 137 |
| Glucose (mmol/l) | 5.4±1.2 | 100 | 5.2±0.8 | 14 | — | — | — | — |
| Insulin (pmol/l) | 157.2±138.0** | 74 | 62.5±41.4 | 14 | — | — | — | — |
Type III HLP: type III hyperlipidemic patients; normo: normolipidemic E2/2 subjects; NDCP: normal Dutch control panel; HTG: hypertriglyceridemic patients. N: total number of subjects with available data about the respective parameter. Body mass index and plasma parameters are presented as mean±SD.
aSelection criterion.
*P<0.05, significantly different from normolipidemic E2/2 subjects.
**P<0.001, significantly different from normolipidemic E2/2 subjects.
Genotype and allele frequencies
Table 2 shows the genotype distributions and allele frequencies of polymorphisms in the APOC3, APOA5 and LPL genes in the type III HLP patients, normolipidemic E2/2 subjects, NDCP and the HTG patients. The genotype or allele frequencies of all HL polymorphisms and a subset of the analyzed LPL polymorphisms did not differ significantly between the study populations and are not shown in Table 2. These include: HL −480 C>T, HL c.219 G>A, HL c.1005 A>G, HL c.609 C>G, LPL c.874 A>G and LPL c.1342 C>G. All polymorphisms, in all groups, were in Hardy–Weinberg equilibrium.
Table 2. Genotype and allele frequencies.
| APOE2 homozygotes | ||||
|---|---|---|---|---|
| Genotype/allele frequencya | Type III HLP, N (%) | NORMO, N (%) | NDCP, N (%) | HTG, N (%) |
| APOC3 3238 G>C | ||||
| GG | 78 (69.6) | 44 (86.3)* | 123 (83.9)** | 71 (61.7) |
| GC | 33 (29.5) | 7 (13.7) | 23 (15.4) | 38 (33.0) |
| CC | 1 (0.9) | 0 (0.0) | 1 (0.7) | 6 (5.2) |
| 1 | 189 (84.4) | 95 (93.1)* | 273 (92.6)* | 180 (78.3) |
| 2 | 35 (15.6) | 7 (6.9) | 25 (7.4) | 50 (21.7) |
| 95 % CI | 1.11–1.20 | 1.02–1.12 | 1.05–1.12 | 1.16–1.27 |
| APOA5 −1131 T>C | ||||
| TT | 75 (70.8) | 45 (88.2)* | 127 (88.2)** | 67 (58.3)* |
| TC | 30 (28.3) | 6 (11.8) | 16 (11.1) | 42 (36.5) |
| CC | 1 (0.9) | 0 (0.0) | 1 (0.7) | 6 (5.2) |
| 1 | 180 (84.9) | 96 (94.1)* | 270 (93.7)** | 176 (76.5)* |
| 2 | 32 (15.1) | 6 (5.9) | 18 (6.3) | 54 (23.5) |
| 95 % CI | 1.10–1.20 | 1.01–1.10 | 1.03–1.09 | 1.18–1.29 |
| APOA5 c.56 G>C | ||||
| GG | 84 (79.2) | 46 (90.2) | 114 (85.1) | 85 (67.5) |
| GC | 19 (17.9) | 5 (9.8) | 20 (14.9) | 35 (27.8) |
| CC | 3 (2.8) | 0 (0.0) | 0 (0.0) | 6 (4.8) |
| 1 | 187 (88.2) | 97 (95.1) | 248 (92.5) | 205 (81.3) |
| 2 | 25 (11.8) | 5 (4.9) | 20 (7.5) | 47 (18.7) |
| 95 % CI | 1.01–1.16 | 1.01–1.10 | 1.04–1.1 | 1.14–1.24 |
| LPL c.27 G>A | ||||
| GG | 100 (88.5) | 51 (98.1)* | 144 (94.7)* | 103 (90.4) |
| GA | 12 (10.6) | 1 (1.9) | 8 (5.3) | 11 (9.6) |
| AA | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 1 | 212 (93.8) | 103 (99.0)* | 296 (97.4)* | 217 (95.2) |
| 2 | 14 (6.2) | 1 (1.0) | 8 (2.6) | 11 (4.8) |
| 95 % CI | 1.03–1.09 | 0.99–1.03 | 1.01–1.04 | 1.02–1.08 |
95% CI: 95% confidence interval; type III HLP: type III hyperlipidemic patients; normo: normolipidemic E2/2 subjects; NDCP: normal Dutch control panel; HTG: hypertriglyceridemic patients.
1 represents common allele frequency; 2 represents rare allele frequency.
*Significant difference genotype/allele frequency (P<0.05) with type III HLP.
**Significant difference genotype/allele frequency (P<0.01) with type III HLP.
Differences between genotype or allele frequencies are based on linear-by-linear association.
Distribution of the genotypes of the APOC3 3238 G>C, APOA5 −1131 T>C and LPL c.27 G>A polymorphisms differed significantly between the normolipidemic E2/2 subjects and the type III HLP patients (Table 2). Distribution of the genotypes of the APOC3 3238 G>C, APOA5 −1131 T>C and LPL c.27 G>A, but not the APOA5 c.56 G>C polymorphisms, differed significantly between NDCP and type III HLP patients. We observed a significant difference in genotype frequency between HTG patients and type III HLP patients for the APOA5 −1131 T>C polymorphisms but we did not observe such difference for the APOC3 3238 G>C, APOA5 c.56 G>C and LPL c.27 G>A polymorphisms.
The frequency of the minor allele of the APOC3 3238 G>C polymorphism and the (in LD) APOA5 −1131 T>C polymorphism was increased in type III HLP patients and the HTG patients (15.6 and 15.1% in the type III HLP, 21.7 and 23.5% in the HTG patients, vs a mean NDCP/normolipidemic APOE2 homozygotes of ∼7.2 and ∼6.1%, respectively). The APOA5 c.56 G>C polymorphism showed a significantly increased frequency of the minor allele in type III HLP patients (11.8%) and in HTG patients (18.7%) when compared with the mean minor allele frequency of the NDCP/normolipidemic APOE2 homozygotes (∼6.2%). A significant enrichment of the minor allele of the LPL c.27 G>A mutation was observed among type III HLP patients (6.2%) as compared to the other study populations (NDCP and normolipidemic APOE2 homozygotes, 2.6 and 1.0%, respectively). LD estimation within the tightly linked cluster of the APOC3/A5 region confirmed earlier findings of others that the APOA5 −1131 T>C and APOC3 3238 G>C polymorphisms are in strong LD (D′=0.8), whereas the APOA5 c.56 G>C and −1131 T>C polymorphisms are not.
Association of APOC3 3238 G>C, APOA5 −1131 T>C, APOA5 c.56 G>C and LPL c.27 G>A with type III HLP
Association between polymorphisms and the expression of type III HLP was tested by logistic regression analysis using a dominant model (ie, 1/1 vs 1/2+2/2). Table 3 shows the association of the APOC3 3238 G>C polymorphism (OR=2.7, 95% CI 1.1–6.7, P=0.03), the APOA5 −1131 T>C polymorphisms (OR=3.1, 95% CI 1.2–8.0, P=0.02), the APOA5 c.56 G>C polymorphism (OR=2.4, 95% CI 0.9–6.7, P=0.12) and the LPL c.27 G>A mutation (OR=6.6, 95% CI 0.8–52.1, P=0.07) with the occurrence of type III HLP.
Table 3. Association of genotypes with the expression of type III HLP.
| Type III HLP, N (%) | Normo, N (%) | OR (95% CI) | P-value | |
|---|---|---|---|---|
| APOC3 3238 G>C | ||||
| 1/1 | 78 (69.6) | 44 (86.3) | 2.7 (1.1–6.7) | 0.03 |
| 1/2+2/2 | 34 (30.4) | 7 (13.7) | ||
| APOA5 −1131 T>C | ||||
| 1/1 | 75 (70.8) | 45 (88.2) | 3.1 (1.2–8.0) | 0.02 |
| 1/2+2/2 | 31 (29.2) | 6 (11.8) | ||
| APOA5 c.56 G>C | ||||
| 1/1 | 84 (79.2) | 46 (90.2) | 2.4 (0.9–6.7) | 0.12 |
| 1/2+2/2 | 22 (20.8) | 5 (9.8) | ||
| LPL c.27 G>A | ||||
| 1/1 | 100 (88.5) | 51 (98.1) | 6.6 (0.8–52.1) | 0.07 |
| 1/2+2/2 | 13 (11.5) | 1 (1.9) | ||
| Cumulative ¥ | ||||
| Non-carrier | 48 (42.5) | 38 (73.1) | 3.7 (1.8–7.5) | <0.0001 |
| Carrier | 65 (57.5) | 14 (26.9) | ||
Type III HLP: type III hyperlipidemic patients; normo: normolipidemic E2/2 subjects; OR: odds ratio; 95% CI: 95% confidence interval; 1/1, homozygous for the common allele; 1/2 heterozygous for the common and rare allele; 2/2, homozygous for the rare allele; cumulative ¥: non-carrier represents non-carrier of the rare allele of either the APOA5 c.56 G>C, SNP3, APOC3 −1131 T>C or LPL c.27 G>A and carrier represents carrier of the rare allele of either the APOA5 c.56 G>C, SNP3, APOC3 −1131 T>C or LPL c.27 G>A.
The APOA5 −1131 T>C and APOC3 3238 G>C polymorphisms (in LD) showed a similar significantly increased risk on the occurrence of type III HLP. A significant association between the APOA5 c.56 G>C polymorphism and the LPL c.27 G>A mutation and expression of type III HLP was not seen. However, the association (Table 3, cumulative) of carrying a rare allele of either APOA5 c.56 G>C, −1131 T>C, APOC3 3238 G>C polymorphisms or LPL c.27 G>A mutation (OR=3.7, 95% CI 1.8–7.5, P<0.0001) and the expression of type III HLP was significantly increased.
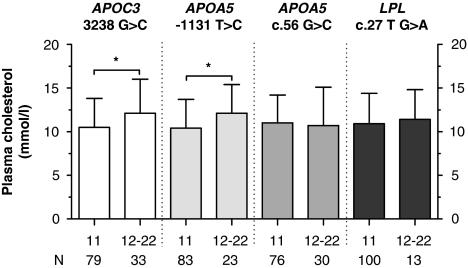
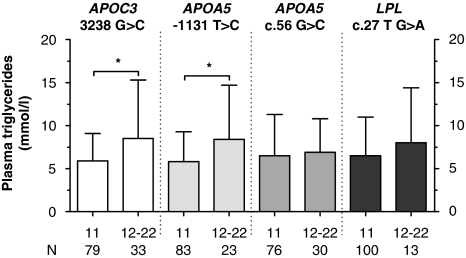
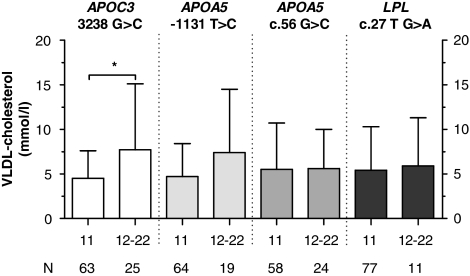
To investigate the effect of the polymorphisms on plasma lipid levels, type III HLP patients were divided into carriers and non-carriers of the polymorphisms. Type III HLP patients with the APOC3 3238 G>C polymorphism and (in LD) the APOA5 −1131 T>C polymorphism showed increased levels of total cholesterol as compared with their counterparts without the polymorphism (Figure 1). Total TG levels were significantly increased in carriers of the APOA5 −1131 T>C polymorphism as compared with non-carriers, whereas the increased levels of total TG levels in the APOC3 3238 G>C carriers did not reach statistical significance (Figure 2). To further determine the association of the different cholesterol-rich lipoprotein fractions with genetic variation, we also investigated the effect of the polymorphisms, APOC3 3238 G>C, APOA5 −1131 T>C and c.56 G>C, and LPL c.27 G>A on plasma HDL cholesterol and VLDL cholesterol (Figure 3). We observed no effect of the polymorphisms on plasma HDL cholesterol (data not shown), whereas carriers of rare variants of both APOC3 3238 G>C and APOA5 −1131 T>C showed a significant association with plasma VLDL cholesterol levels. Evaluation of the association of the APOA5 c.56 G>C, APOA5 −1131 T>C, APOC3 3238 G>C and the LPL c.27 G>A variants with plasma lipid levels in the NDCP did not reveal any significant associations. The effect of the polymorphisms on lipid levels in the HTG patients has been described earlier.23, 24
Figure 1.
Effect of the APOC3 3238 G>C, the APOA5 −1131 T>C and APOA5 c.56 G>C and the LPL c.27 G>A polymorphisms on plasma cholesterol levels in type III HLP patients. N represents number of type III HLP patients. *P<0.05.
Figure 2.
Effect of the APOC3 3238 G>C, the APOA5 −1131 T>C and APOA5 c.56 G>C, and the LPL c.27 G>A polymorphisms on plasma triglyceride levels in type III HLP patients. N represents number of type III HLP patients. *P<0.05.
Figure 3.
Effect of the APOC3 3238 G>C, the APOA5 −1131 T>C and APOA5 c.56 G>C, and the LPL c.27 G>A polymorphisms on VLDL-cholesterol levels in type III HLP patients. N represents number of type III HLP patients. *P<0.05.
Discussion
The clinical manifestation of type III HLP, in addition to defective hepatic clearance of remnant lipoproteins in E2/2 subjects, may be caused by impaired lipolysis.4, 9, 40 Several groups reported about LPL and proteins involved in LPL activity, as apoCIII and apoAV in type III patients.13, 14, 15, 16 In accordance with this hypothesis, we replicated in the largest type III cohort thus far, associations for polymorphisms in proteins involved in the lipolysis of lipoproteins: the 3238 G>C polymorphism in the APOC3 gene (in LD with APOA5 −1131 T>C polymorphism) and the c.27 G>A mutation in the LPL gene. Our results show that the APOC3 3238 G>C/APOA5 −1131 T>C polymorphism exacerbates the hyperlipidemic phenotype of type III HLP patients, whereas the LPL c.27 G>A mutation has no additional effect on plasma lipid levels.
In an earlier study, we found an interaction between hyperinsulinemia and the APOC3 3238 G>C polymorphism, associated with severe hyperlipidemia in APOE2 homozygotes.12 By increasing the number of E2/2 subjects, data from the present study demonstrate that the APOC3 3238 G>C polymorphism per se is also an important contributor to type III HLP expression. The APOC3 3238 G>C polymorphism is strongly associated with elevated plasma TG levels.41 In accordance, our type III HLP patients carrying the APOC3 3238 G>C polymorphism showed a more severe hyperlipidemia, ie higher VLDL cholesterol levels and a tendency of higher TG levels than non-carriers. The molecular mechanism underlying the association with HTG is still unclear. The APOC3 3238 G>C polymorphism is located in the 3′-UTR of the APOC3 gene and could be a causal variant by acting on mRNA stability, which results in increasing apoCIII plasma levels. However, it is also possible that the APOC3 3238 G>C polymorphism itself is not responsible for the TG-raising effect. Other mutations within the APOA1-C3-A4 gene cluster, located near the APOC3 3238 G>C polymorphic site, could be candidates for being the causative mutations leading to HTG. Two polymorphisms located at positions −482 and −455 in the promoter region of the APOC3 gene showed substantial LD with the 3238 G>C polymorphism, as reported by others.41, 42, 43 However, we found in a much smaller subgroup of our type III HLP patients, no evidence of an association between these polymorphisms and increased TG levels,12 whereas Miller et al40 suggested an association with these two APOC3 polymorphisms and the metabolic syndrome. Functional studies to elucidate the exact role on apoCIII protein levels of each of these three polymorphisms should be performed.
The LPL c.27 G>A mutation leads to an increased bridging of LDL and monocyte adhesion and is also associated with elevated levels of plasma TGs and reduced plasma HDL cholesterol levels.44 In our type III HLP patients, a tendency toward increased plasma cholesterol and TG levels was observed in LPL c.27 G>A carriers, but this did not reach statistical significance, possibly due to the size of our groups. In Caucasians, the –93T → G transition in the LPL gene promoter is in LD with the LPL c.27 G>A mutation.18, 45 However, several studies have reported that the TG-raising effect was solely attributable to the presence of the LPL c.27 G>A mutation.45
Data from genetic association studies on type III HLP are limited due to the low prevalence of APOE2 homozygosity in the population. To circumvent this problem, most studies have compared the allele frequency of mutations in type III HLP patients with that of the general population. Zhang et al13 found an increased allele frequency for the LPL N291S mutation in type III HLP patients as compared with the Dutch population. However, this approach does not include the possibility that the mutation is also more prevalent in normolipidemic APOE2 homozygotes. Conversely, Evans and Beil16 showed that the LPL mutations D9N, in particular N291S and S447X (c.27 G>A, c.874 A>G and c.1342 C>G, respectively), were not associated with type III HLP.
Comparison of the Dutch population frequency for the HL −480 C>T mutation (19%) with that of our type III HLP patients (31%) revealed that the mutation was more prevalent in patients (P<0.001).36 As we did not find such a difference between patients and normolipidemic E2/2 subjects, we may conclude that the LPL c.874 A>G and the HL −480 C>T mutations are not major contributors to the expression of type III HLP (data not shown). In addition, no association with type III HLP was found for HL c.219 G>A, 1005 A>G, 609 C>G and LPL c.1342 C>G.
To further study the patient and population prevalences of the APOA5, APOC3 polymorphisms and the LPL c.27 G>A mutation, two additional populations were screened: the NDCP and HTG patients. The prevalences of the APOC3 3238 G>C/APOA5 −1131 T>C polymorphism, APOA5 c.56 G>C and the LPL c.27 G>A mutation in the NDCP cohort are similar to those observed in the normolipidemic APOE2 homozygote individuals. This indicates that the normolipidemic APOE2 homozygotes were not a selected sub-population. In contrast, the prevalences of the APOC3, APOAV and LPL mutations in the HTG patients are similar to those of the type III patients, indicating that these polymorphisms are universal risk factors for HTG.
Earlier studies have reported frequencies for the rare variant of the c.56 G>C polymorphism in type III patients.14, 15, 46 In our type III patients, the rare variant of the APOA5 c.56 G>C polymorphism is far less frequent (11.8%) than reported thus far. In addition, 4.9% of the normolipidemic APOE2 homozygotes still was carrier of the rare allele of the APOA5 c.56 G>C polymorphism. Moreover, we found no association between plasma TG and cholesterol levels and the c.56 G>C polymorphism in hyper- and normolipidemic FDs. The fact that this association is not present and the frequency of the c.56 C-allele is somewhat increased in the type III HLP population suggests that the APOA5 c.56 G>C polymorphism is a weak contributor to the clinical manifestation of type III HLP. The mechanism by which this variant acts remains to be solved. It has been suggested that the APOA5 c.56 G>C polymorphism alters the signal peptide, which results in a decreased secretion of apoAV protein. However, such correlation between APOA5 rare variant carriers and plasma apoAV was not found in a cohort of HTG patients.23
As reported by others, our data show that the APOC3 3238 G>C and APOA5 −1131 T>C polymorphisms show a high LD.41, 47 In total, 15% of type III HLP patients are carriers of the APOA5 −1131 T>C polymorphism and 15.6% are carriers of the APOC3 3238 G>C polymorphism. In the present study, we did not aim to elucidate a causal role of each polymorphism.
In total, 58% of type III HLP patients vs 27% of normolipidemic APOE2 homozygotes are carriers of the APOC3 3238 G>C/APOA5 −1131 T>C polymorphism, APOA5 c.56 G>C and/or the LPL c.27 G>A mutation, indicating that these mutations could partly explain the expression of type III HLP. Additional genetic and environmental risk factors remain to be identified. Earlier, we found that hyperinsulinemic APOE2 homozygotes have an increased risk for type III HLP.6 It is likely that the suppression of VLDL production by insulin in these hyperinsulinemic patients is reduced, resulting in chronically elevated VLDL production.10 Another possible mechanism is through an interaction between insulin and genes involved in the lipolysis of lipoproteins. Several studies support the hypothesis that insulin is involved in the regulation of the APOC3 gene. The presence of the APOC3 –455 and –482 polymorphisms may abolish the insulin responsiveness of the APOC3 promoter.48, 49 As the APOC3 3238 G>C polymorphism in E2/2 subjects is almost exclusively found in combination with the presence of the –455 and –482 promoter variants, it seems possible that in E2/2 subjects, who are carriers of the 3238 G>C polymorphism, the loss of insulin regulation results in overexpression of the APOC3 gene and, as a consequence, in overt hyperlipidemia.12, 34 In addition, insulin is a major regulator of LPL activity. The combination of high insulin levels and decreased LPL activity, as found in carriers of LPL mutations, may further exacerbate the expression of HTG.50
In conclusion, our data indicate that the APOC3 3238 G>C, APOA5 −1131 T>C and, to a lesser extent, LPL c.27 G>A mutation associate with a more severe hyperlipidemia in type III patients, whereas APOA5 c.56 G>C is perhaps a weak modifier.
Acknowledgments
This study was supported by the Netherlands Heart Foundation (project no. 94.114) and by the Nutrigenomics Consortium. Sylvia Kamerling, Leonie van Vark and Ton Vroom are thanked for expert technical assistance. The Rotterdam Study is supported by the Netherlands Organisation for Scientific Research and the Municipality of Rotterdam. We thank Jeannette Vergeer, Wilma Luijten and Bianca de Graaf for their help in the laboratory analysis of the Rotterdam Study.
References
- Morganroth J, Levy RI, Fredrickson DS. The biochemical, clinical, and genetic features of type III hyperlipoproteinemia. Ann Intern Med. 1975;82:158–174. doi: 10.7326/0003-4819-82-2-158. [DOI] [PubMed] [Google Scholar]
- Brewer HB, Jr, Zech LA, Gregg RE, Schwartz D, Schaefer EJ. NIH Conference. Type III hyperlipoproteinemia: diagnosis, molecular defects, pathology, and treatment. Ann Intern Med. 1983;98:623–640. doi: 10.7326/0003-4819-98-5-623. [DOI] [PubMed] [Google Scholar]
- Smelt AH, de Beer F. Apolipoprotein E and familial dysbetalipoproteinemia: clinical, biochemical, and genetic aspects. Semin Vasc Med. 2004;4:249–257. doi: 10.1055/s-2004-861492. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Utermann G. Apolipoprotein E polymorphism in health and disease. Am Heart J. 1987;113:433–440. doi: 10.1016/0002-8703(87)90610-7. [DOI] [PubMed] [Google Scholar]
- de Beer F, Stalenhoef AF, Hoogerbrugge N, et al. Expression of type III hyperlipoproteinemia in apolipoprotein E2 (Arg158 → Cys) homozygotes is associated with hyperinsulinemia. Arterioscler Thromb Vasc Biol. 2002;22:294–299. doi: 10.1161/hq0202.102919. [DOI] [PubMed] [Google Scholar]
- Utermann G, Hees M, Steinmetz A. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604–607. doi: 10.1038/269604a0. [DOI] [PubMed] [Google Scholar]
- Zhao SP, Smelt AH, Van den Maagdenberg AM, et al. Plasma lipoprotein profiles of normocholesterolemic and hypercholesterolemic homozygotes for apolipoprotein E2 (Arg158 → Cys) compared. Clin Chem. 1994;40:1559–1566. [PubMed] [Google Scholar]
- Mahley RW, Huang Y, Rall SC., Jr Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- Olefsky JM, Farquhar JW, Reaven GM. Reappraisal of the role of insulin in hypertriglyceridemia. Am J Med. 1974;57:551–560. doi: 10.1016/0002-9343(74)90006-0. [DOI] [PubMed] [Google Scholar]
- Zammit VA, Waterman IJ, Topping D, McKay G. Insulin stimulation of hepatic triacylglycerol secretion and the etiology of insulin resistance. J Nutr. 2001;131:2074–2077. doi: 10.1093/jn/131.8.2074. [DOI] [PubMed] [Google Scholar]
- Sijbrands EJ, Hoffer MJ, Meinders AE, et al. Severe hyperlipidemia in apolipoprotein E2 homozygotes due to a combined effect of hyperinsulinemia and an SstI polymorphism. Arterioscler Thromb Vasc Biol. 1999;19:2722–2729. doi: 10.1161/01.atv.19.11.2722. [DOI] [PubMed] [Google Scholar]
- Zhang H, Reymer PW, Liu MS, et al. Patients with apoE3 deficiency (E2/2, E3/2, and E4/2) who manifest with hyperlipidemia have increased frequency of an Asn 291 → Ser mutation in the human LPL gene. Arterioscler Thromb Vasc Biol. 1995;15:1695–1703. doi: 10.1161/01.atv.15.10.1695. [DOI] [PubMed] [Google Scholar]
- Schaefer JR, Sattler AM, Hackler B, et al. Hyperlipidemia in patients with apolipoprotein E2/2 phenotype: apolipoprotein A5 S19W mutation as a cofactor. Clin Chem. 2004;50:2214. doi: 10.1373/clinchem.2004.037689. [DOI] [PubMed] [Google Scholar]
- Evans D, Seedorf U, Beil FU. Polymorphisms in the apolipoprotein A5 (APOA5) gene and type III hyperlipidemia. Clin Genet. 2005;68:369–372. doi: 10.1111/j.1399-0004.2005.00510.x. [DOI] [PubMed] [Google Scholar]
- Evans D, Beil FU. The D9N, N291S and S447X variants in the lipoprotein lipase (LPL) gene are not associated with type III hyperlipidemia. BMC Med Genet. 2007;8:56. doi: 10.1186/1471-2350-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud PJ, Humphries SE. Genetic polymorphisms, lipoproteins and coronary artery disease risk. Curr Opin Lipidol. 2001;12:405–409. doi: 10.1097/00041433-200108000-00006. [DOI] [PubMed] [Google Scholar]
- Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- Jansen H, Verhoeven AJ, Sijbrands EJ. Hepatic lipase: a pro- or anti-atherogenic protein. J Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200. [DOI] [PubMed] [Google Scholar]
- Zambon A, Deeb SS, Pauletto P, Crepaldi G, Brunzell JD. Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. Curr Opin Lipidol. 2003;14:179–189. doi: 10.1097/00041433-200304000-00010. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- O'Brien PJ, Alborn WE, Sloan JH, et al. The novel apolipoprotein A5 is present in human serum, is associated with VLDL, HDL, and chylomicrons, and circulates at very low concentrations compared with other apolipoproteins. Clin Chem. 2005;51:351–359. doi: 10.1373/clinchem.2004.040824. [DOI] [PubMed] [Google Scholar]
- Henneman P, Schaap FG, Havekes LM, et al. Plasma apoAV levels are markedly elevated in severe hypertriglyceridemia and positively correlated with the APOA5 S19W polymorphism. Atherosclerosis. 2007;193:129–134. doi: 10.1016/j.atherosclerosis.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Henneman P, Schaap FG, Rensen PC, van Dijk KW, Smelt AH. Estrogen induced hypertriglyceridemia in an apolipoprotein AV deficient patient. J Intern Med. 2008;263:107–108. doi: 10.1111/j.1365-2796.2007.01889.x. [DOI] [PubMed] [Google Scholar]
- Assman G, Schulte H.Lipid Metabolism Disorders and Coronary Heart Disease. Results and Conclusions of the Prospective Cardiovascular Munster (PROCAM) Study2nd Eds. München: MMV Medizin Verlag. 199319–67.
- Smit M, De Knijff P, Rosseneu M, et al. Apolipoprotein E polymorphism in The Netherlands and its effect on plasma lipid and apolipoprotein levels. Hum Genet. 1988;80:287–292. doi: 10.1007/BF01790099. [DOI] [PubMed] [Google Scholar]
- Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- Stolk RP, Pols HA, Lamberts SW, de Jong PT, Hofman A, Grobbee DE. Diabetes mellitus, impaired glucose tolerance, and hyperinsulinemia in an elderly population. The Rotterdam Study. Am J Epidemiol. 1997;145:24–32. doi: 10.1093/oxfordjournals.aje.a009028. [DOI] [PubMed] [Google Scholar]
- Havekes LM, De Knijff P, Beisiegel U, Havinga J, Smit M, Klasen E. A rapid micromethod for apolipoprotein E phenotyping directly in serum. J Lipid Res. 1987;28:455–463. [PubMed] [Google Scholar]
- Reymer PW, Groenemeyer BE, van de BR, Kastelein JJ. Apolipoprotein E genotyping on agarose gels. Clin Chem. 1995;41:1046–1047. [PubMed] [Google Scholar]
- Burstein M, Scholnick HR, Morfin R. Rapid method for the isolation of lipoproteins from human serum by precipitation with polyanions. J Lipid Res. 1970;11:583–595. [PubMed] [Google Scholar]
- Starr JI, Mako ME, Juhn D, Rubenstein AH. Measurement of serum proinsulin-like material: cross-reactivity of porcine and human proinsulin in the insulin radioimmunoassay. J Lab Clin Med. 1978;91:683–692. [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer MJ, Sijbrands EJ, De Man FH, Havekes LM, Smelt AH, Frants RR. Increased risk for endogenous hypertriglyceridaemia is associated with an apolipoprotein C3 haplotype specified by the SstI polymorphism. Eur J Clin Invest. 1998;28:807–812. doi: 10.1046/j.1365-2362.1998.00361.x. [DOI] [PubMed] [Google Scholar]
- Hoffer MJ, Bredie SJ, Snieder H, et al. Gender-related association between the −93T → G/D9N haplotype of the lipoprotein lipase gene and elevated lipid levels in familial combined hyperlipidemia. Atherosclerosis. 1998;138:91–99. doi: 10.1016/s0021-9150(98)00007-0. [DOI] [PubMed] [Google Scholar]
- Jansen H, Verhoeven AJ, Weeks L, et al. Common C-to-T substitution at position −480 of the hepatic lipase promoter associated with a lowered lipase activity in coronary artery disease patients. Arterioscler Thromb Vasc Biol. 1997;17:2837–2842. doi: 10.1161/01.atv.17.11.2837. [DOI] [PubMed] [Google Scholar]
- Knudsen P, Antikainen M, Ehnholm S, et al. A compound heterozygote for hepatic lipase gene mutations Leu334 → Phe and Thr383 → Met: correlation between hepatic lipase activity and phenotypic expression. J Lipid Res. 1996;37:825–834. [PubMed] [Google Scholar]
- Hegele RA, Tu L, Connelly PW. Human hepatic lipase mutations and polymorphisms. Hum Mutat. 1992;1:320–324. doi: 10.1002/humu.1380010410. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Miller M, Rhyne J, Chen H, et al. APOC3 promoter polymorphisms C-482T and T-455C are associated with the metabolic syndrome. Arch Med Res. 2007;38:444–451. doi: 10.1016/j.arcmed.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud PJ, Hawe E, Martin S, et al. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum Mol Genet. 2002;11:3039–3046. doi: 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3′ untranslated region polymorphisms. Proc Natl Acad Sci USA. 1993;90:4562–4566. doi: 10.1073/pnas.90.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CQ, Parnell LD, Ordovas JM. The APOA1/C3/A4/A5 gene cluster, lipid metabolism and cardiovascular disease risk. Curr Opin Lipidol. 2005;16:153–166. doi: 10.1097/01.mol.0000162320.54795.68. [DOI] [PubMed] [Google Scholar]
- Fisher RM, Benhizia F, Schreiber R, et al. Enhanced bridging function and augmented monocyte adhesion by lipoprotein lipase N9: insights into increased risk of coronary artery disease in N9 carriers. Atherosclerosis. 2003;166:243–251. doi: 10.1016/s0021-9150(02)00337-4. [DOI] [PubMed] [Google Scholar]
- Talmud PJ, Stephens JW. Lipoprotein lipase gene variants and the effect of environmental factors on cardiovascular disease risk. Diabetes Obes Metab. 2004;6:1–7. doi: 10.1111/j.1463-1326.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- Martin-Campos JM, Rico N, Bonet R, Mayoral C, Ordonez-Llanos J, Blanco-Vaca F. Apolipoprotein A5 S19W may play a role in dysbetalipoproteinemia in patients with the Apo E2/E2 genotype. Clin Chem. 2006;52:1974–1975. doi: 10.1373/clinchem.2006.070318. [DOI] [PubMed] [Google Scholar]
- Olivier M, Wang X, Cole R, et al. Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics. 2004;83:912–923. doi: 10.1016/j.ygeno.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Breslow JL, Li W, Leff T. Transcriptional regulation of the apoC-III gene by insulin in diabetic mice: correlation with changes in plasma triglyceride levels. J Lipid Res. 1994;35:1918–1924. [PubMed] [Google Scholar]
- Altomonte J, Cong L, Harbaran S, et al. Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest. 2004;114:1493–1503. doi: 10.1172/JCI19992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Vohl MC, Gaudet D, et al. Hyperinsulinemia and abdominal obesity affect the expression of hypertriglyceridemia in heterozygous familial lipoprotein lipase deficiency. Diabetes. 1997;46:2063–2068. doi: 10.2337/diab.46.12.2063. [DOI] [PubMed] [Google Scholar]